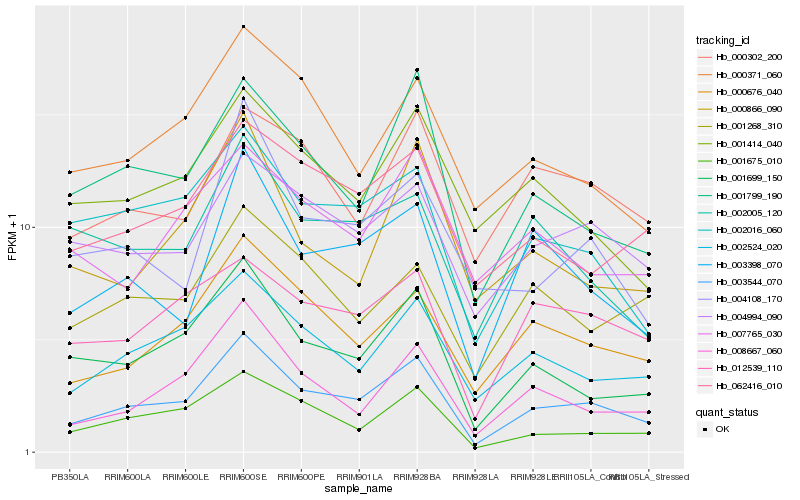
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003544_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001699_150 |
0.1108499521 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 3 |
Hb_000676_040 |
0.1150618618 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 4 |
Hb_001675_010 |
0.115358949 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 5 |
Hb_002524_020 |
0.1156129349 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 6 |
Hb_001799_190 |
0.1190118312 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 7 |
Hb_008667_060 |
0.1233688041 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 8 |
Hb_003398_070 |
0.1239339133 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 9 |
Hb_002016_060 |
0.1259683033 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 10 |
Hb_012539_110 |
0.1308376814 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_004994_090 |
0.1316395916 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 12 |
Hb_062416_010 |
0.13447341 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 13 |
Hb_000371_060 |
0.1353041198 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 14 |
Hb_000302_200 |
0.1413294184 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 15 |
Hb_001414_040 |
0.1422101882 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000866_090 |
0.1438748684 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_004108_170 |
0.1457548193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002005_120 |
0.1462945394 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 19 |
Hb_001268_310 |
0.1464494974 |
- |
- |
- |
| 20 |
Hb_007765_030 |
0.1472919569 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |