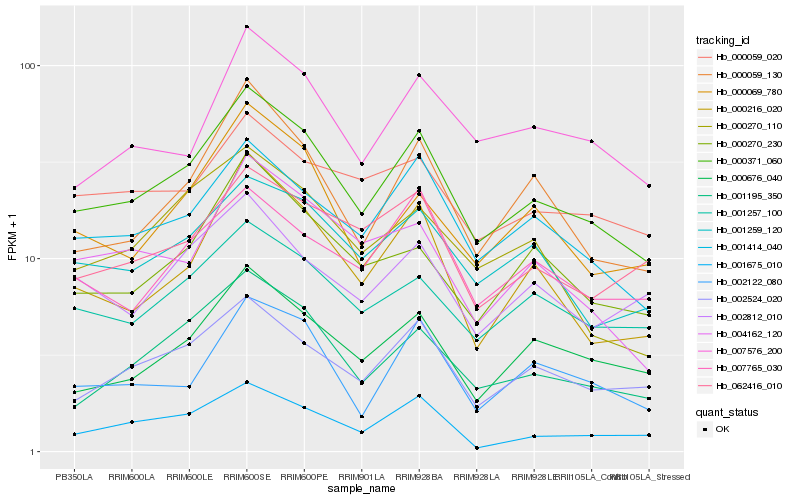
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000371_060 |
0.0 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 2 |
Hb_001414_040 |
0.08124374 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002524_020 |
0.0867383668 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 4 |
Hb_001259_120 |
0.0920023102 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 5 |
Hb_001195_350 |
0.0921254894 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 6 |
Hb_001257_100 |
0.0932932692 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 7 |
Hb_000059_130 |
0.0962531007 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000676_040 |
0.0966046629 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 9 |
Hb_000216_020 |
0.1031332829 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 10 |
Hb_000270_230 |
0.1032453839 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 11 |
Hb_000069_780 |
0.1036586427 |
- |
- |
atpob1, putative [Ricinus communis] |
| 12 |
Hb_007576_200 |
0.1051752999 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_004162_120 |
0.1096214802 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 14 |
Hb_007765_030 |
0.1113092702 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 15 |
Hb_062416_010 |
0.1125464889 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 16 |
Hb_000270_110 |
0.1133712959 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 17 |
Hb_000059_020 |
0.1134070918 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 18 |
Hb_002122_080 |
0.1148470964 |
- |
- |
- |
| 19 |
Hb_001675_010 |
0.1149431099 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 20 |
Hb_002812_010 |
0.1154489874 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |