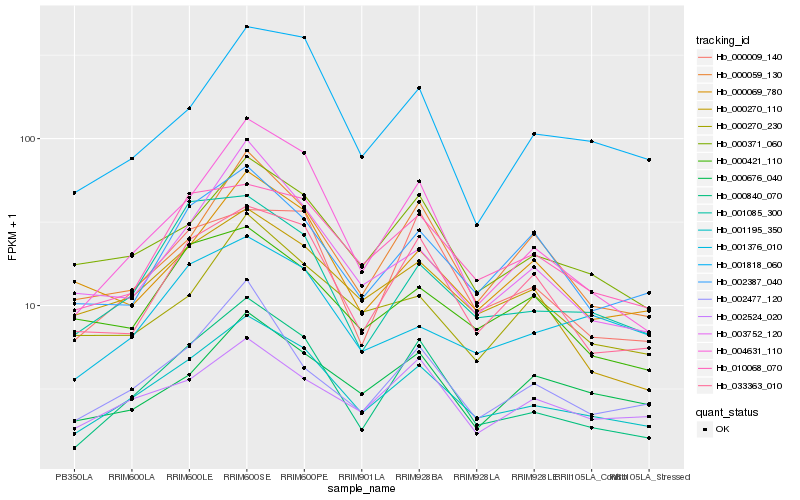
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_350 |
0.0 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 2 |
Hb_000371_060 |
0.0921254894 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 3 |
Hb_010068_070 |
0.1050420241 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_004631_110 |
0.1056679507 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_000840_070 |
0.1065003812 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_033363_010 |
0.1084619208 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 7 |
Hb_002524_020 |
0.1125121325 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_000270_110 |
0.1143530781 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 9 |
Hb_000069_780 |
0.1144778467 |
- |
- |
atpob1, putative [Ricinus communis] |
| 10 |
Hb_002387_040 |
0.1150512586 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 11 |
Hb_000059_130 |
0.1156454463 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 12 |
Hb_001085_300 |
0.1187265825 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_000676_040 |
0.1207949611 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 14 |
Hb_001818_060 |
0.1210914686 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 15 |
Hb_001376_010 |
0.1215536636 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 16 |
Hb_000270_230 |
0.1217520459 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 17 |
Hb_002477_120 |
0.1228410858 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_000009_140 |
0.1236843632 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 19 |
Hb_003752_120 |
0.1250932382 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000421_110 |
0.125777759 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |