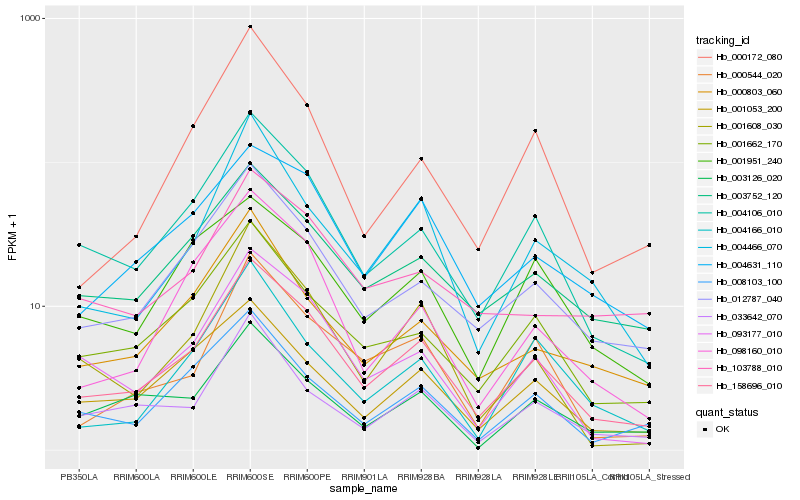
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158696_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 2 |
Hb_004106_010 |
0.0835815629 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012787_040 |
0.0980958908 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001662_170 |
0.1063348574 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_003752_120 |
0.111712583 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 6 |
Hb_093177_010 |
0.1151330887 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 7 |
Hb_004631_110 |
0.1186218868 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_004466_070 |
0.1247616308 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 9 |
Hb_001053_200 |
0.125622381 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 10 |
Hb_000544_020 |
0.1273288012 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 11 |
Hb_008103_100 |
0.1295928259 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 12 |
Hb_000803_060 |
0.1309732276 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 13 |
Hb_001608_030 |
0.1315255826 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 14 |
Hb_098160_010 |
0.1336302012 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_033642_070 |
0.1349318086 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 16 |
Hb_000172_080 |
0.1369879207 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 17 |
Hb_003126_020 |
0.1424064145 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 18 |
Hb_001951_240 |
0.1444826291 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 19 |
Hb_004166_010 |
0.1487509035 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 20 |
Hb_103788_010 |
0.1489821388 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |