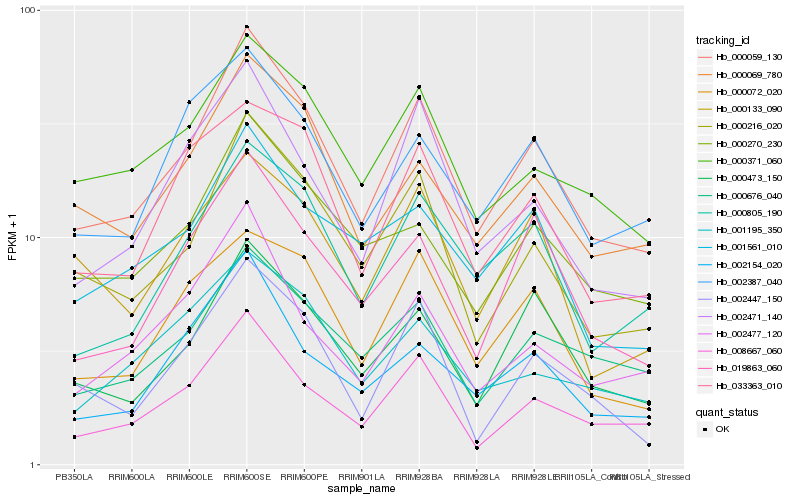
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_130 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000216_020 |
0.0799712977 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 3 |
Hb_008667_060 |
0.0882107462 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 4 |
Hb_000473_150 |
0.0936571967 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000805_190 |
0.0955586296 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 6 |
Hb_000069_780 |
0.0957137926 |
- |
- |
atpob1, putative [Ricinus communis] |
| 7 |
Hb_000371_060 |
0.0962531007 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 8 |
Hb_000676_040 |
0.1002744767 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 9 |
Hb_019863_060 |
0.1017179392 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 10 |
Hb_000270_230 |
0.1032269463 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 11 |
Hb_002471_140 |
0.1049792143 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 12 |
Hb_033363_010 |
0.1054297679 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 13 |
Hb_001561_010 |
0.1063254287 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000072_020 |
0.1065398173 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002154_020 |
0.107822413 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_002387_040 |
0.1078937445 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 17 |
Hb_001195_350 |
0.1156454463 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 18 |
Hb_002477_120 |
0.1162164102 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_002447_150 |
0.1181499556 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_000133_090 |
0.1199369024 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |