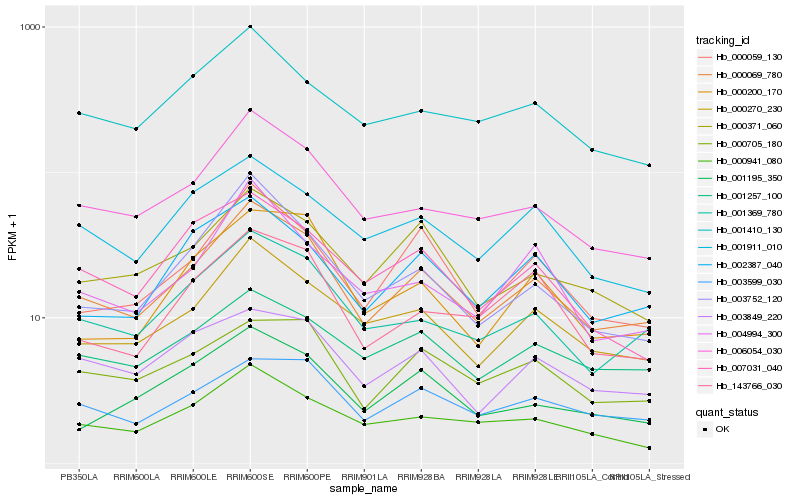
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_780 |
0.0 |
- |
- |
atpob1, putative [Ricinus communis] |
| 2 |
Hb_000270_230 |
0.0789806854 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 3 |
Hb_006054_030 |
0.0821304316 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 4 |
Hb_001369_780 |
0.0833250134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000059_130 |
0.0957137926 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 6 |
Hb_002387_040 |
0.1000489688 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_003752_120 |
0.1001657297 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000941_080 |
0.1020977548 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 9 |
Hb_000371_060 |
0.1036586427 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 10 |
Hb_001257_100 |
0.1037258205 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_001410_130 |
0.1058070208 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 12 |
Hb_003599_030 |
0.1063277656 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_001911_010 |
0.1066024618 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 14 |
Hb_003849_220 |
0.1110327831 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 15 |
Hb_143766_030 |
0.1115377749 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000705_180 |
0.1124007834 |
- |
- |
hypothetical protein JCGZ_02923 [Jatropha curcas] |
| 17 |
Hb_004994_300 |
0.1130345594 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 18 |
Hb_000200_170 |
0.1143315518 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 19 |
Hb_001195_350 |
0.1144778467 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 20 |
Hb_007031_040 |
0.1150941121 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |