| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
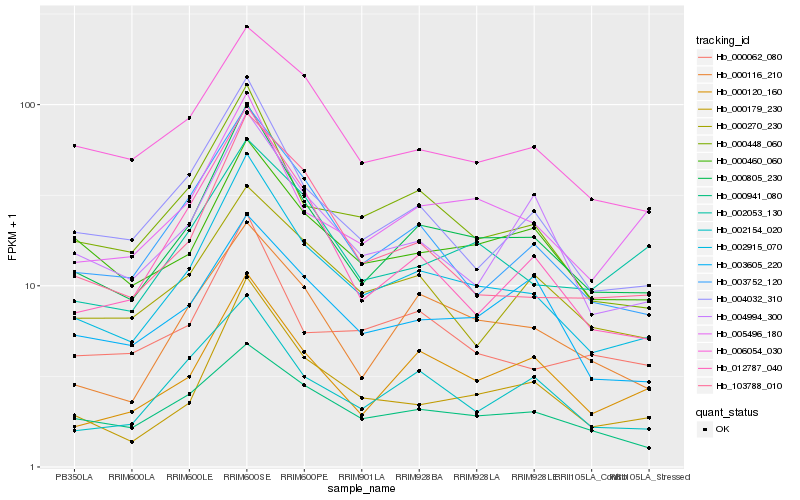
Hb_002915_070 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 2 |
Hb_000805_230 |
0.0515973249 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 3 |
Hb_000448_060 |
0.0855103482 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 4 |
Hb_000179_230 |
0.0964282148 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 5 |
Hb_003752_120 |
0.0983068915 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004032_310 |
0.1005572698 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_103788_010 |
0.1149895748 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_002154_020 |
0.1156821599 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000116_210 |
0.1181977738 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 10 |
Hb_006054_030 |
0.1189261413 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 11 |
Hb_004994_300 |
0.1195011975 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 12 |
Hb_000062_080 |
0.1197929456 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 13 |
Hb_005496_180 |
0.1201719009 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 14 |
Hb_000120_160 |
0.1236648196 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000941_080 |
0.1242759651 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 16 |
Hb_000460_060 |
0.1246017395 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 17 |
Hb_002053_130 |
0.1305293249 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 18 |
Hb_012787_040 |
0.1307393704 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000270_230 |
0.1311027251 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 20 |
Hb_003605_220 |
0.1315632187 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |