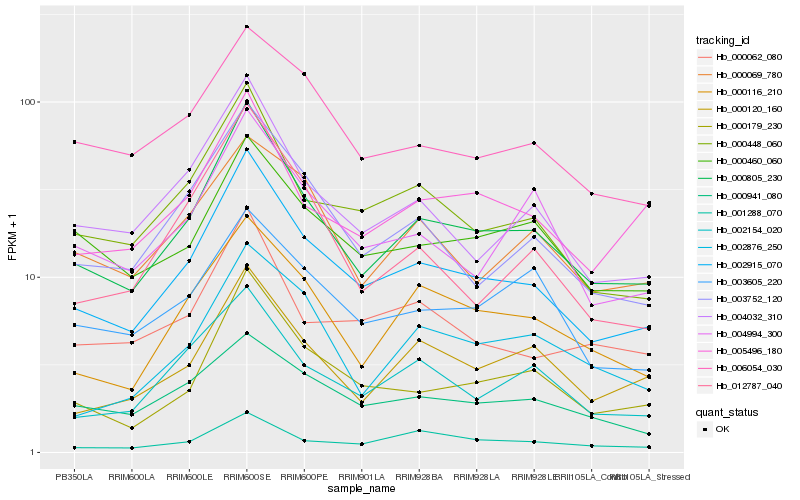
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_230 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 2 |
Hb_002915_070 |
0.0515973249 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 3 |
Hb_000179_230 |
0.1002404989 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 4 |
Hb_000120_160 |
0.1020205287 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000116_210 |
0.1030404449 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 6 |
Hb_004032_310 |
0.103918066 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_000448_060 |
0.1062789865 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 8 |
Hb_003752_120 |
0.1078246509 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002154_020 |
0.1142674812 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_005496_180 |
0.1161717226 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 11 |
Hb_004994_300 |
0.1183425267 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 12 |
Hb_002876_250 |
0.1221077003 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 13 |
Hb_000460_060 |
0.1221682478 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 14 |
Hb_006054_030 |
0.1268753285 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 15 |
Hb_000069_780 |
0.1291787412 |
- |
- |
atpob1, putative [Ricinus communis] |
| 16 |
Hb_003605_220 |
0.1297585412 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 17 |
Hb_000062_080 |
0.1307748953 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 18 |
Hb_001288_070 |
0.1308803237 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 19 |
Hb_000941_080 |
0.1311635544 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 20 |
Hb_012787_040 |
0.1312898454 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |