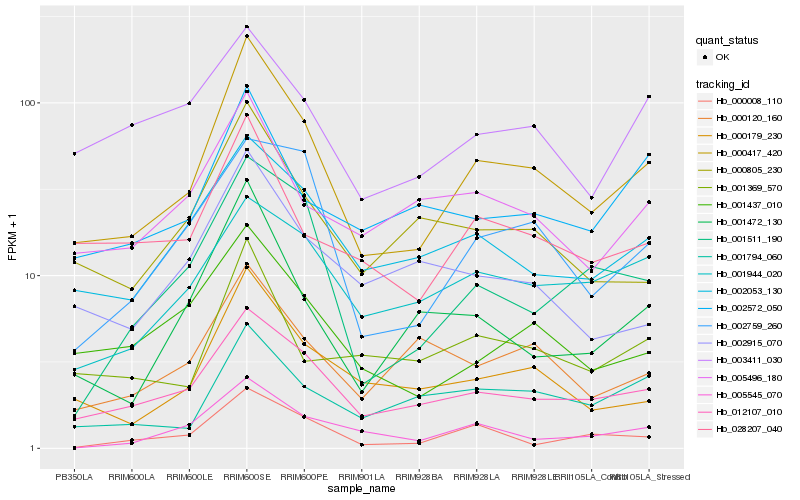
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_420 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 2 |
Hb_012107_010 |
0.1191908261 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 3 |
Hb_000179_230 |
0.1423183898 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 4 |
Hb_002053_130 |
0.1543717077 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 5 |
Hb_001437_010 |
0.1596805869 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 6 |
Hb_028207_040 |
0.1622241088 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 7 |
Hb_001472_130 |
0.1629370589 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 8 |
Hb_000805_230 |
0.1656182997 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 9 |
Hb_001511_190 |
0.1689630974 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 10 |
Hb_002759_260 |
0.1692729419 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 11 |
Hb_005545_070 |
0.1745411749 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_001794_060 |
0.1748085653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001369_570 |
0.176349744 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 14 |
Hb_005496_180 |
0.1775832352 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 15 |
Hb_000008_110 |
0.1793195401 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 16 |
Hb_002572_050 |
0.1793874267 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_000120_160 |
0.1797963861 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001944_020 |
0.1808497083 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 19 |
Hb_002915_070 |
0.1858645912 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 20 |
Hb_003411_030 |
0.186305644 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |