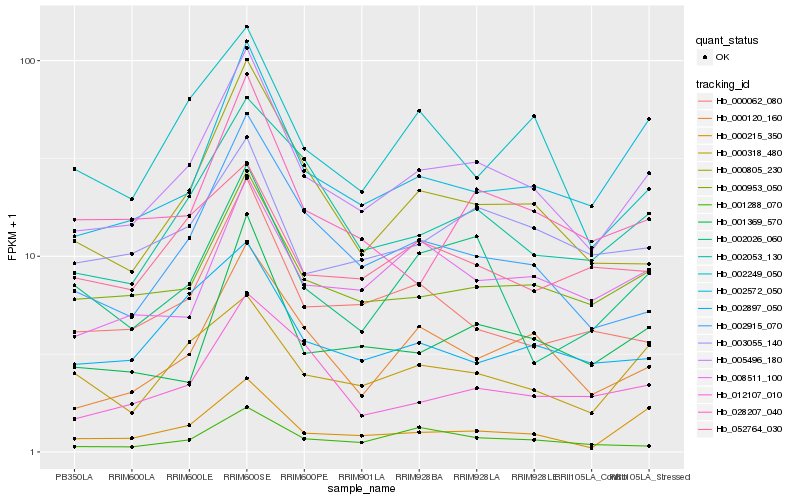
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005496_180 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 2 |
Hb_000953_050 |
0.1018277226 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 3 |
Hb_002572_050 |
0.1158996767 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000805_230 |
0.1161717226 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 5 |
Hb_000120_160 |
0.1162924905 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002053_130 |
0.1200914151 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 7 |
Hb_002915_070 |
0.1201719009 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 8 |
Hb_001369_570 |
0.1235974687 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 9 |
Hb_000062_080 |
0.1277448146 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 10 |
Hb_003055_140 |
0.1298230553 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 11 |
Hb_002897_050 |
0.1298345321 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 12 |
Hb_008511_100 |
0.1312230567 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 13 |
Hb_000215_350 |
0.1331178302 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_028207_040 |
0.1339120615 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 15 |
Hb_002026_060 |
0.1361804485 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 16 |
Hb_000318_480 |
0.1361984737 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 17 |
Hb_001288_070 |
0.1378404435 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 18 |
Hb_002249_050 |
0.1405328655 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 19 |
Hb_012107_010 |
0.1434464322 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 20 |
Hb_052764_030 |
0.1438674111 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |