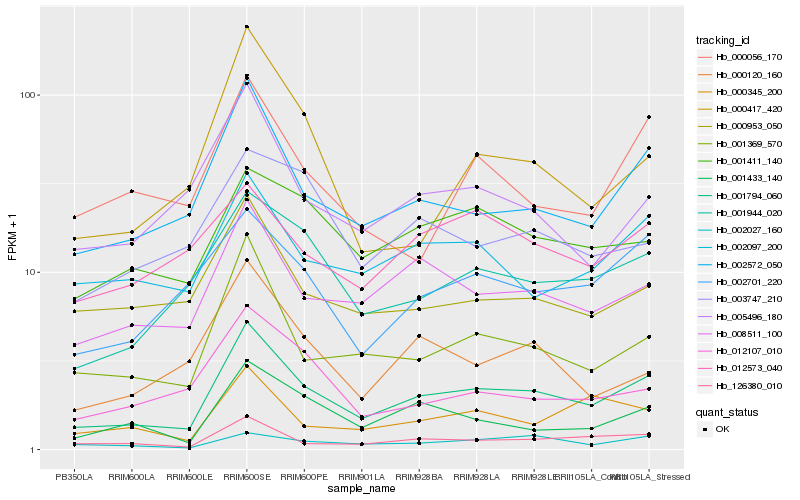
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001794_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002572_050 |
0.1182148031 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_126380_010 |
0.130423498 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_001369_570 |
0.1305567631 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 5 |
Hb_008511_100 |
0.1338952048 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 6 |
Hb_001944_020 |
0.1412773388 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 7 |
Hb_001433_140 |
0.1473145364 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 8 |
Hb_002701_220 |
0.154577498 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 9 |
Hb_002097_200 |
0.1547709456 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_000953_050 |
0.1600699985 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 11 |
Hb_001411_140 |
0.1615883423 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 12 |
Hb_005496_180 |
0.1634699952 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 13 |
Hb_000120_160 |
0.1646146737 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000345_200 |
0.1671170967 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 15 |
Hb_012107_010 |
0.1678596632 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 16 |
Hb_002027_160 |
0.1700874069 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_000056_170 |
0.172735162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000417_420 |
0.1748085653 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 19 |
Hb_003747_210 |
0.1781155104 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 20 |
Hb_012573_040 |
0.1791671953 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |