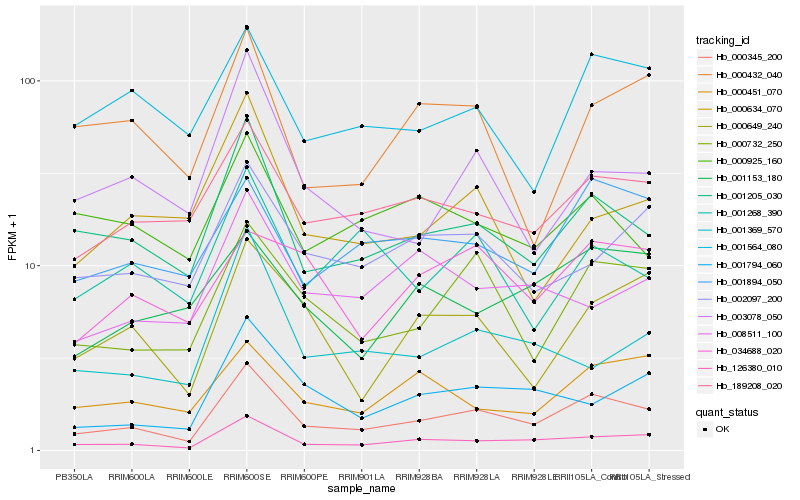
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 2 |
Hb_126380_010 |
0.0939963404 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001205_030 |
0.1230703881 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 4 |
Hb_000732_250 |
0.1359484296 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 5 |
Hb_189208_020 |
0.1450422958 |
- |
- |
LRX2, putative [Ricinus communis] |
| 6 |
Hb_001894_050 |
0.1585083974 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 7 |
Hb_001794_060 |
0.1671170967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001564_080 |
0.1680363478 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_000649_240 |
0.1703301999 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 10 |
Hb_002097_200 |
0.1706529308 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 11 |
Hb_000451_070 |
0.1750051694 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_003078_050 |
0.1771311164 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 13 |
Hb_001153_180 |
0.1789642911 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 14 |
Hb_000432_040 |
0.1818571441 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 15 |
Hb_001369_570 |
0.1820770274 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 16 |
Hb_034688_020 |
0.1832188868 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 17 |
Hb_001268_390 |
0.1841297454 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 18 |
Hb_008511_100 |
0.1848074067 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 19 |
Hb_000634_070 |
0.1851937964 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 20 |
Hb_000925_160 |
0.1883851865 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |