| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
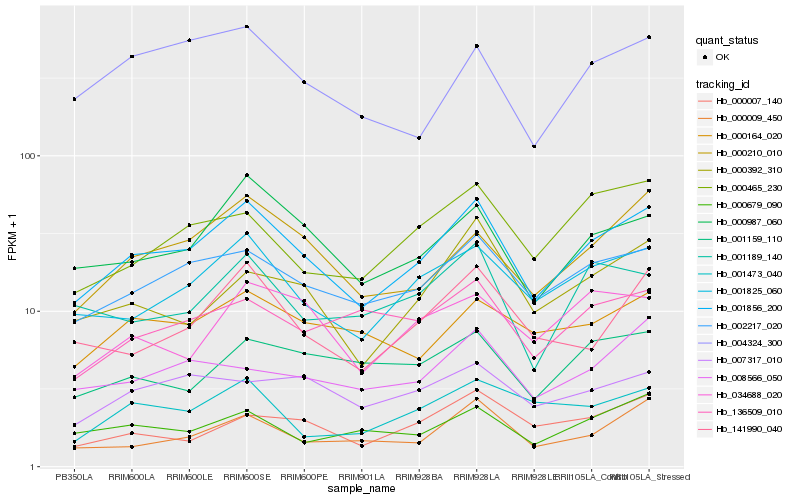
Hb_001856_200 |
0.0 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 2 |
Hb_002217_020 |
0.0930377879 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001825_060 |
0.0948786995 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 4 |
Hb_000987_060 |
0.1016307886 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 5 |
Hb_000210_010 |
0.1111336491 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 6 |
Hb_141990_040 |
0.1200807229 |
- |
- |
Protein SCAR2, putative [Ricinus communis] |
| 7 |
Hb_000009_450 |
0.1216971841 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000164_020 |
0.1297464819 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000465_230 |
0.1305938387 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 10 |
Hb_000392_310 |
0.1311492868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000679_090 |
0.1323219596 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 12 |
Hb_004324_300 |
0.1336948888 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 13 |
Hb_000007_140 |
0.1344379601 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 14 |
Hb_034688_020 |
0.1351715605 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 15 |
Hb_001189_140 |
0.1357185321 |
- |
- |
- |
| 16 |
Hb_001473_040 |
0.1364436907 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_008566_050 |
0.1371485558 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 18 |
Hb_001159_110 |
0.1373280981 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 19 |
Hb_136509_010 |
0.1373755533 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 20 |
Hb_007317_010 |
0.1389631321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |