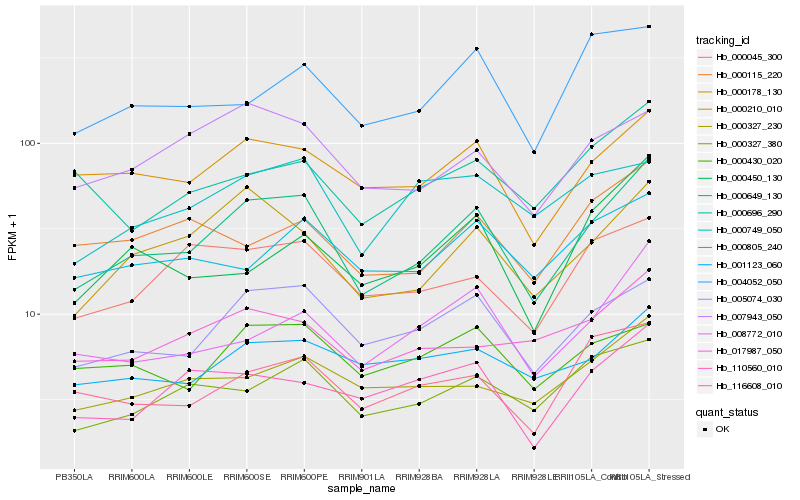
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_130 |
0.0 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 2 |
Hb_005074_030 |
0.0995683246 |
- |
- |
- |
| 3 |
Hb_000210_010 |
0.1063107884 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 4 |
Hb_000178_130 |
0.1147963982 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 5 |
Hb_000327_380 |
0.1245459997 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 6 |
Hb_116608_010 |
0.1307082814 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_007943_050 |
0.1308351982 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 8 |
Hb_000327_230 |
0.1310279076 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008772_010 |
0.1314773804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001123_060 |
0.1327538307 |
- |
- |
PREDICTED: ALBINO3-like protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000045_300 |
0.1329994757 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 12 |
Hb_017987_050 |
0.1349813475 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000430_020 |
0.1375423394 |
- |
- |
- |
| 14 |
Hb_110560_010 |
0.1398576829 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000805_240 |
0.1409360437 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 16 |
Hb_000696_290 |
0.1414793096 |
- |
- |
PREDICTED: UPF0664 stress-induced protein C29B12.11c [Jatropha curcas] |
| 17 |
Hb_000115_220 |
0.1417969096 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 18 |
Hb_004052_050 |
0.141919439 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |
| 19 |
Hb_000450_130 |
0.1445479652 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 20 |
Hb_000749_050 |
0.1447354991 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |