| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
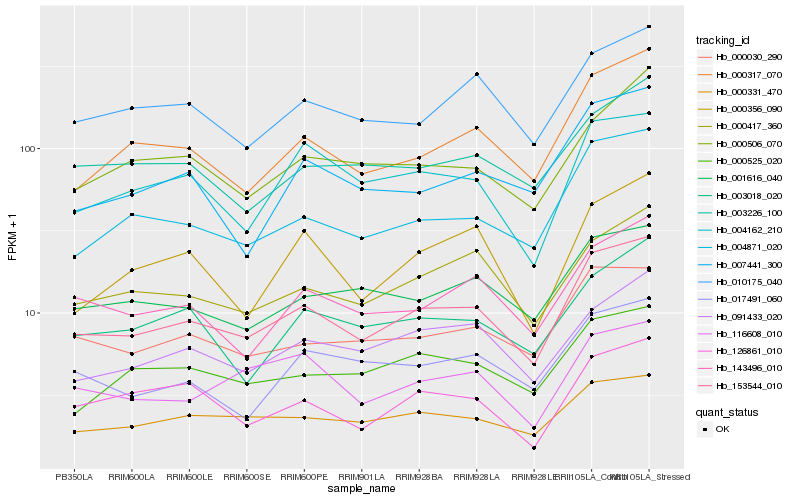
Hb_153544_010 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 2 |
Hb_004871_020 |
0.0691499893 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 3 |
Hb_000030_290 |
0.0792042572 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 4 |
Hb_000317_070 |
0.0795206676 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 5 |
Hb_000417_360 |
0.0830686761 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_091433_020 |
0.0859735664 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_143496_010 |
0.0864342158 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3 [Glycine max] |
| 8 |
Hb_010175_040 |
0.0877695655 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6a, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001616_040 |
0.0891134904 |
- |
- |
PREDICTED: uncharacterized protein LOC105644362 [Jatropha curcas] |
| 10 |
Hb_000331_470 |
0.0910949639 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 11 |
Hb_000525_020 |
0.0944042702 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_003018_020 |
0.0949862946 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |
| 13 |
Hb_003226_100 |
0.0952321351 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 14 |
Hb_126861_010 |
0.0979664198 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 15 |
Hb_007441_300 |
0.0984427123 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_017491_060 |
0.0985387493 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000506_070 |
0.0989388601 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 18 |
Hb_004162_210 |
0.0992456469 |
- |
- |
Cyclin-dependent kinases regulatory subunit, putative [Ricinus communis] |
| 19 |
Hb_000356_090 |
0.1000937306 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 20 |
Hb_116608_010 |
0.1006437988 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |