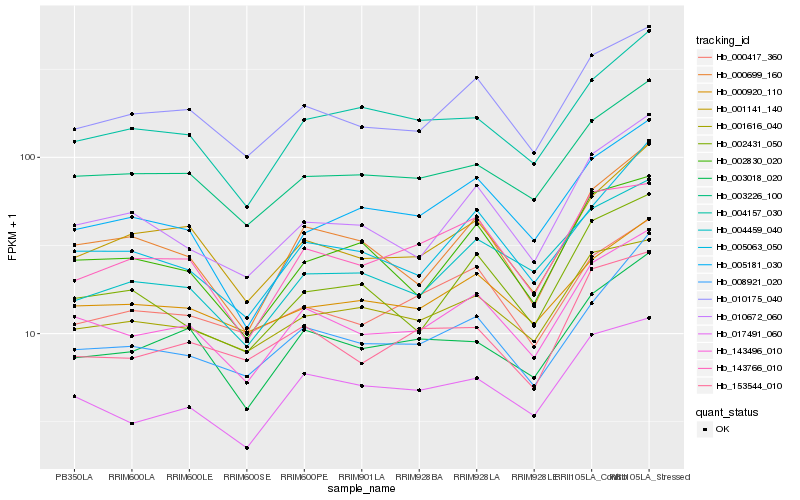
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143496_010 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3 [Glycine max] |
| 2 |
Hb_010175_040 |
0.0572180548 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6a, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003226_100 |
0.0633747733 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 4 |
Hb_017491_060 |
0.0699678663 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001141_140 |
0.0717861702 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |
| 6 |
Hb_003018_020 |
0.0729271013 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |
| 7 |
Hb_000699_160 |
0.0729606037 |
- |
- |
PREDICTED: C-Myc-binding protein homolog [Jatropha curcas] |
| 8 |
Hb_005063_050 |
0.0745224796 |
- |
- |
PREDICTED: probable prefoldin subunit 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010672_060 |
0.0765687154 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 10 |
Hb_000920_110 |
0.0776031184 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP4 [Jatropha curcas] |
| 11 |
Hb_000417_360 |
0.0796027693 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001616_040 |
0.0797006946 |
- |
- |
PREDICTED: uncharacterized protein LOC105644362 [Jatropha curcas] |
| 13 |
Hb_002830_020 |
0.0817827691 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_008921_020 |
0.0824604235 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 15 |
Hb_004157_030 |
0.0839976413 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 16 |
Hb_004459_040 |
0.0846021509 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 17 |
Hb_005181_030 |
0.0847542019 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 18 |
Hb_002431_050 |
0.0861875826 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 [Jatropha curcas] |
| 19 |
Hb_143766_010 |
0.0862156018 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 20 |
Hb_153544_010 |
0.0864342158 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |