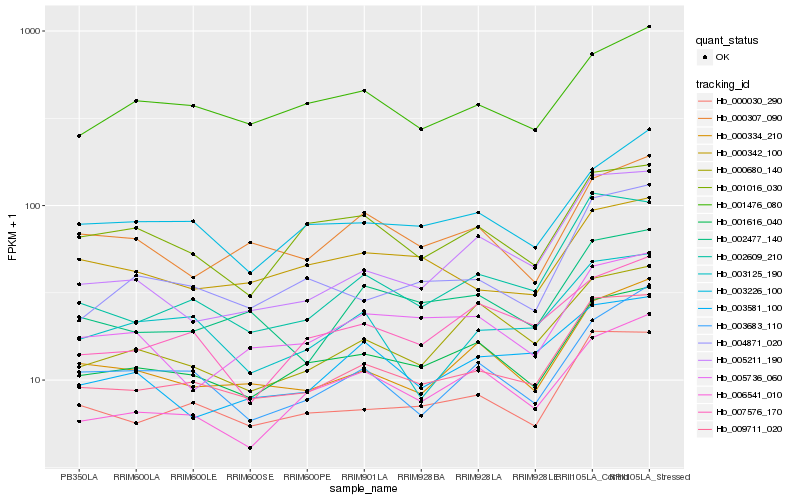
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009711_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647794 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000030_290 |
0.04919048 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 3 |
Hb_002609_210 |
0.0582238432 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_001616_040 |
0.0644111652 |
- |
- |
PREDICTED: uncharacterized protein LOC105644362 [Jatropha curcas] |
| 5 |
Hb_000334_210 |
0.0766422109 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002477_140 |
0.0775620538 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004871_020 |
0.0858009964 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 8 |
Hb_001476_080 |
0.0889668546 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5A-like [Jatropha curcas] |
| 9 |
Hb_003226_100 |
0.0912019406 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 10 |
Hb_000680_140 |
0.0915722902 |
- |
- |
PREDICTED: uncharacterized protein LOC105631472 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006541_010 |
0.0921173198 |
- |
- |
PREDICTED: uncharacterized protein LOC105637261 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003581_100 |
0.0931026802 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005736_060 |
0.0945296694 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 14 |
Hb_005211_190 |
0.0947009153 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 15 |
Hb_000307_090 |
0.0949729985 |
- |
- |
hypothetical protein CICLE_v10002475mg [Citrus clementina] |
| 16 |
Hb_001016_030 |
0.0978696102 |
- |
- |
PREDICTED: uncharacterized protein LOC105636529 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003683_110 |
0.0981028755 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 18 |
Hb_007576_170 |
0.0989822032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003125_190 |
0.1000428647 |
- |
- |
hypothetical protein POPTR_0015s10960g [Populus trichocarpa] |
| 20 |
Hb_000342_100 |
0.1007751558 |
- |
- |
PREDICTED: PITH domain-containing protein 1 [Jatropha curcas] |