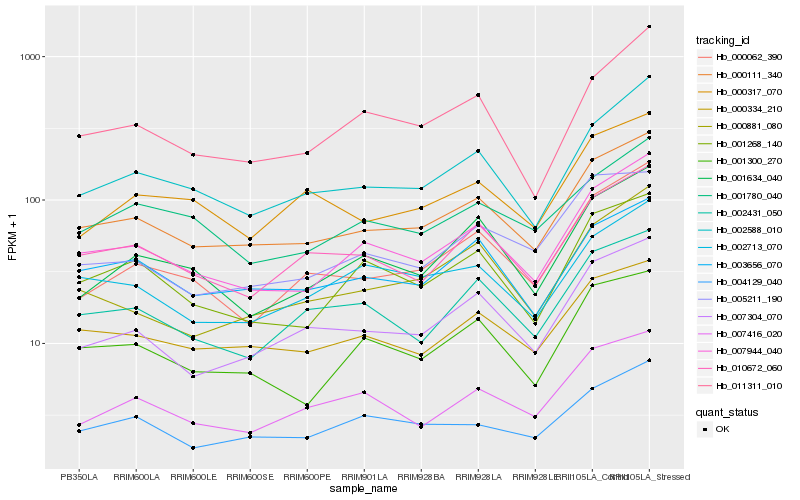
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_340 |
0.0 |
- |
- |
peptidyl-prolyl cis-trans isomerase h, ppih, putative [Ricinus communis] |
| 2 |
Hb_007944_040 |
0.0551645476 |
- |
- |
60S ribosomal protein L18aB [Hevea brasiliensis] |
| 3 |
Hb_000334_210 |
0.0602261536 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010672_060 |
0.0661899674 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 5 |
Hb_002588_010 |
0.0674601911 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 6 |
Hb_007304_070 |
0.0695860088 |
- |
- |
hypoxanthine-guanine phosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003656_070 |
0.0734206566 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 8 |
Hb_004129_040 |
0.0788638157 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Vitis vinifera] |
| 9 |
Hb_001300_270 |
0.081083198 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 10 |
Hb_002431_050 |
0.0814196819 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 [Jatropha curcas] |
| 11 |
Hb_001268_140 |
0.0823817358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002713_070 |
0.0841316104 |
- |
- |
hypothetical protein POPTR_0002s21620g [Populus trichocarpa] |
| 13 |
Hb_007416_020 |
0.08527388 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Cucumis sativus] |
| 14 |
Hb_000062_390 |
0.0875272496 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 15 |
Hb_005211_190 |
0.0878907751 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 16 |
Hb_011311_010 |
0.0884462028 |
- |
- |
60S ribosomal protein L27C [Hevea brasiliensis] |
| 17 |
Hb_000881_080 |
0.0896389407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000317_070 |
0.0899297095 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 19 |
Hb_001780_040 |
0.0901584064 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 20 |
Hb_001634_040 |
0.0902791158 |
transcription factor |
TF Family: NF-YC |
DNA polymerase epsilon subunit, putative [Ricinus communis] |