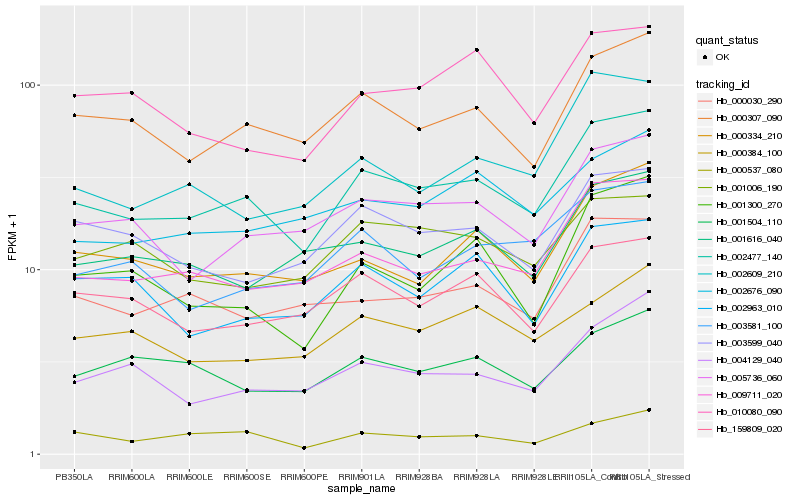
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_140 |
0.0 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000307_090 |
0.0761306959 |
- |
- |
hypothetical protein CICLE_v10002475mg [Citrus clementina] |
| 3 |
Hb_009711_020 |
0.0775620538 |
- |
- |
PREDICTED: uncharacterized protein LOC105647794 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005736_060 |
0.0839348533 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 5 |
Hb_000334_210 |
0.0948124208 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000030_290 |
0.0951848476 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 7 |
Hb_003599_040 |
0.0990486012 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_001616_040 |
0.0992859194 |
- |
- |
PREDICTED: uncharacterized protein LOC105644362 [Jatropha curcas] |
| 9 |
Hb_159809_020 |
0.099900405 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000537_080 |
0.101352474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002676_090 |
0.101718939 |
- |
- |
PREDICTED: protein TIC 22-like, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003581_100 |
0.1029232198 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004129_040 |
0.1057459252 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Vitis vinifera] |
| 14 |
Hb_002609_210 |
0.1057830848 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000384_100 |
0.1067314516 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 16 |
Hb_002963_010 |
0.1072946186 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_010080_090 |
0.1087702785 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 [Jatropha curcas] |
| 18 |
Hb_001504_110 |
0.1091493056 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |
| 19 |
Hb_001300_270 |
0.1094579391 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 20 |
Hb_001006_190 |
0.1109083481 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |