| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
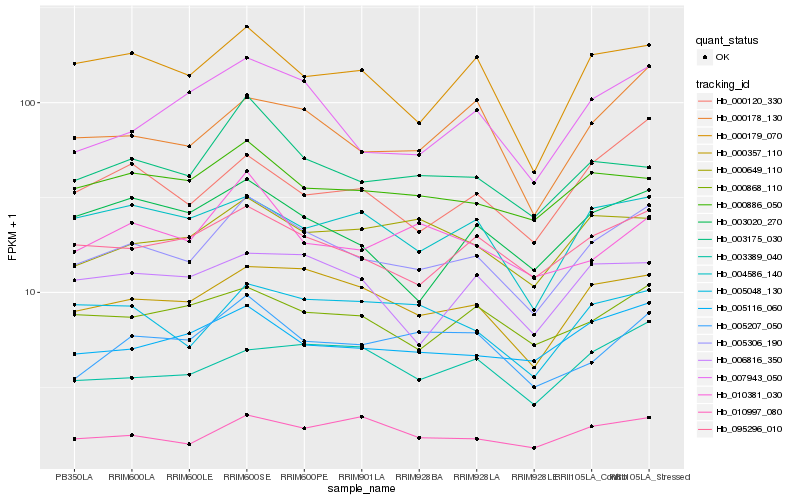
Hb_005306_190 |
0.0 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 2 |
Hb_000357_110 |
0.0736395601 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 3 |
Hb_095296_010 |
0.0791331328 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 4 |
Hb_005116_060 |
0.0879098553 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007943_050 |
0.0887213592 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 6 |
Hb_000178_130 |
0.0890807921 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 7 |
Hb_000179_070 |
0.0939147197 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 8 |
Hb_003175_030 |
0.0948232854 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 9 |
Hb_000868_110 |
0.0949172675 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 10 |
Hb_005207_050 |
0.0950372957 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 39 [Jatropha curcas] |
| 11 |
Hb_000120_330 |
0.0970139876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000886_050 |
0.0980489182 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 13 |
Hb_010997_080 |
0.0993325808 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_004586_140 |
0.0994485034 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 15 |
Hb_005048_130 |
0.099749009 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 16 |
Hb_003389_040 |
0.0999111489 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 17 |
Hb_000649_110 |
0.1019547015 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 18 |
Hb_003020_270 |
0.1026121009 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 19 |
Hb_010381_030 |
0.1033181334 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 20 |
Hb_006816_350 |
0.1051759164 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |