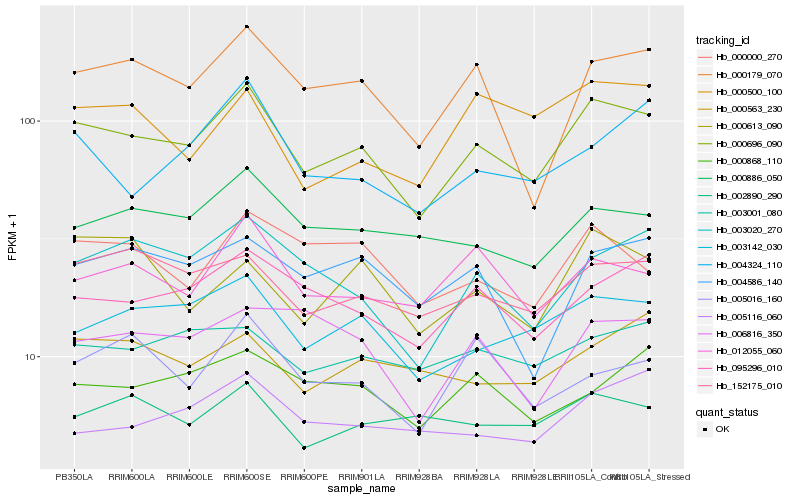
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 2 |
Hb_003142_030 |
0.0772795513 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 3 |
Hb_152175_010 |
0.082878272 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 4 |
Hb_095296_010 |
0.0850291296 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 5 |
Hb_003020_270 |
0.0879581242 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 6 |
Hb_012055_060 |
0.0899921205 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000179_070 |
0.0905964135 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 8 |
Hb_000886_050 |
0.0936023814 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 9 |
Hb_000000_270 |
0.0956615905 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185 [Jatropha curcas] |
| 10 |
Hb_003001_080 |
0.0963484277 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 11 |
Hb_004324_110 |
0.0973718863 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 12 |
Hb_000868_110 |
0.0996326658 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 13 |
Hb_005116_060 |
0.1014389498 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004586_140 |
0.1017080434 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 15 |
Hb_000613_090 |
0.1017333146 |
- |
- |
PREDICTED: uncharacterized protein LOC105641542 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000563_230 |
0.1023911574 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 17 |
Hb_000500_100 |
0.1025865713 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 18 |
Hb_005016_160 |
0.1026823012 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 19 |
Hb_002890_290 |
0.1037506145 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 20 |
Hb_006816_350 |
0.1039744151 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |