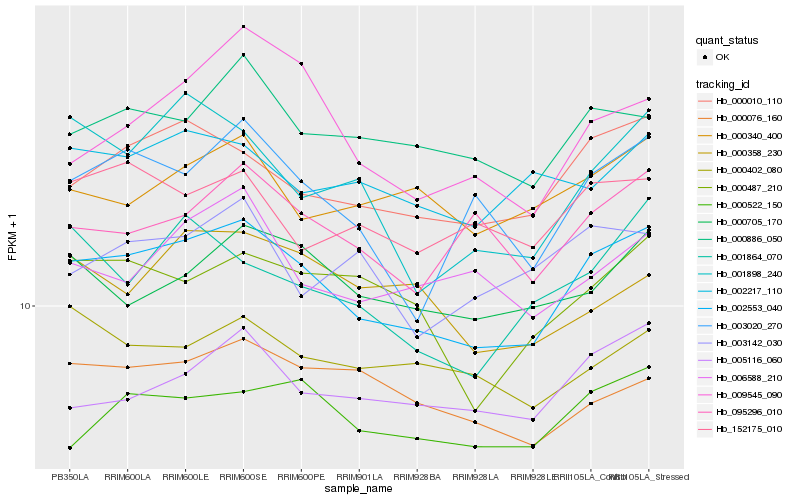
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002553_040 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 2 |
Hb_000010_110 |
0.0721555538 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003020_270 |
0.0862186676 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 4 |
Hb_000886_050 |
0.0864719469 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 5 |
Hb_000522_150 |
0.0877341605 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 6 |
Hb_000487_210 |
0.0891804891 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 7 |
Hb_005116_060 |
0.0898026404 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000705_170 |
0.0907371688 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 9 |
Hb_095296_010 |
0.0918960943 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 10 |
Hb_152175_010 |
0.0944665628 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 11 |
Hb_000076_160 |
0.095130978 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 12 |
Hb_000340_400 |
0.0958732743 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 13 |
Hb_002217_110 |
0.0966609573 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 14 |
Hb_006588_210 |
0.0969957288 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 15 |
Hb_000402_080 |
0.0976564845 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 16 |
Hb_001898_240 |
0.098049299 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 17 |
Hb_001864_070 |
0.0998636257 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 18 |
Hb_009545_090 |
0.1002651777 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 19 |
Hb_000358_230 |
0.1013812808 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_003142_030 |
0.1015284914 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |