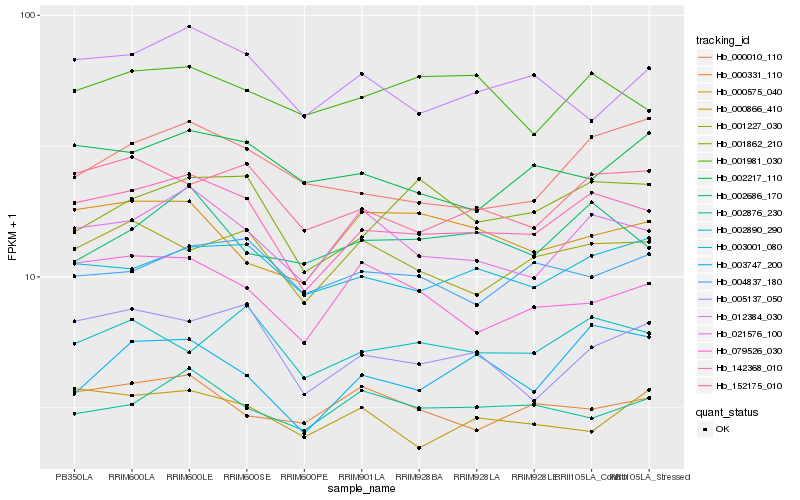
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142368_010 |
0.0 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_021576_100 |
0.0665533799 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_152175_010 |
0.0708087424 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 4 |
Hb_003001_080 |
0.0760900846 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 5 |
Hb_001227_030 |
0.0767538969 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005137_050 |
0.0827821814 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 7 |
Hb_001862_210 |
0.0845706442 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 8 |
Hb_002890_290 |
0.0858729184 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 9 |
Hb_079526_030 |
0.0862306111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 10 |
Hb_002686_170 |
0.0901939498 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012384_030 |
0.0906959477 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 12 |
Hb_001981_030 |
0.0909340229 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000331_110 |
0.0912498632 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 14 |
Hb_002217_110 |
0.0914753397 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 15 |
Hb_004837_180 |
0.0917554228 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003747_200 |
0.0930093346 |
- |
- |
PREDICTED: DNA cross-link repair protein SNM1 [Jatropha curcas] |
| 17 |
Hb_000010_110 |
0.0934090253 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000575_040 |
0.0952203837 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 19 |
Hb_000866_410 |
0.0953296934 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 20 |
Hb_002876_230 |
0.0954363052 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |