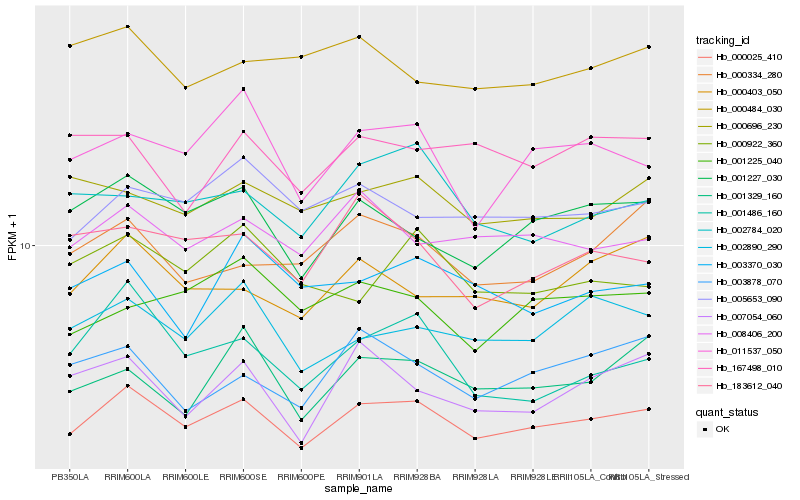
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_410 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 2 |
Hb_001227_030 |
0.0614747969 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008406_200 |
0.0625326559 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002890_290 |
0.0664715677 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005653_090 |
0.0666141369 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 6 |
Hb_000696_230 |
0.0699517 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 7 |
Hb_000403_050 |
0.0708301123 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_011537_050 |
0.0709104411 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002784_020 |
0.0747758117 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_001329_160 |
0.0748017088 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000334_280 |
0.0758830158 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 12 |
Hb_003878_070 |
0.0764127586 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 13 |
Hb_183612_040 |
0.0765233499 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 14 |
Hb_001225_040 |
0.0767433499 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 15 |
Hb_167498_010 |
0.0770734177 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 16 |
Hb_003370_030 |
0.0774021801 |
- |
- |
PREDICTED: uncharacterized protein LOC105635547 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001486_160 |
0.0777182726 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 18 |
Hb_000484_030 |
0.0779614636 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 19 |
Hb_007054_060 |
0.0779707538 |
- |
- |
- |
| 20 |
Hb_000922_360 |
0.0785762656 |
- |
- |
- |