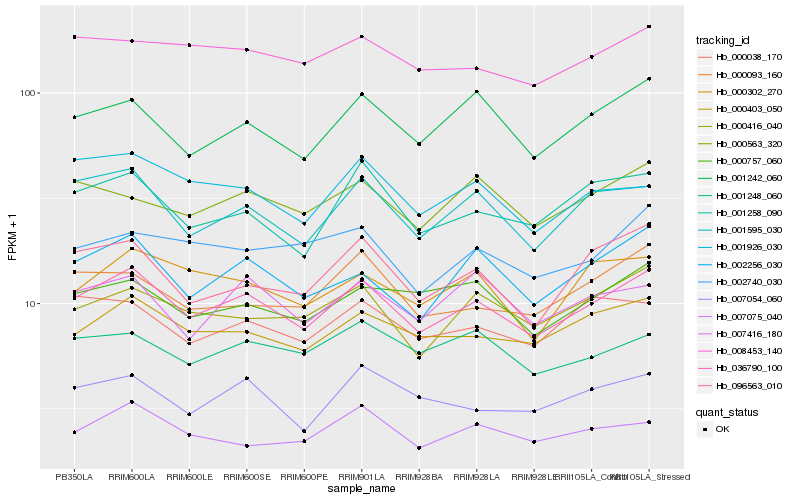
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036790_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |
| 2 |
Hb_001595_030 |
0.0562813123 |
- |
- |
PREDICTED: nuclear inhibitor of protein phosphatase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000403_050 |
0.058711007 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001242_060 |
0.0612815145 |
- |
- |
PREDICTED: uncharacterized protein LOC105139508 [Populus euphratica] |
| 5 |
Hb_002256_030 |
0.0642841936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001258_090 |
0.0660159845 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 7 |
Hb_000416_040 |
0.0679147304 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 8 |
Hb_096563_010 |
0.0693540882 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 9 |
Hb_000757_060 |
0.0713135197 |
- |
- |
hypothetical protein PRUPE_ppa001141mg [Prunus persica] |
| 10 |
Hb_007054_060 |
0.0718528131 |
- |
- |
- |
| 11 |
Hb_008453_140 |
0.0721359255 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 12 |
Hb_000038_170 |
0.0724471421 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 13 |
Hb_000302_270 |
0.072608726 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001248_060 |
0.0726131558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000563_320 |
0.0729006253 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 16 |
Hb_007075_040 |
0.0731530201 |
- |
- |
hypothetical protein CISIN_1g039518mg, partial [Citrus sinensis] |
| 17 |
Hb_007416_180 |
0.0732336024 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 18 |
Hb_000093_160 |
0.074165338 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002740_030 |
0.0741817727 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001926_030 |
0.0745867106 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |