| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
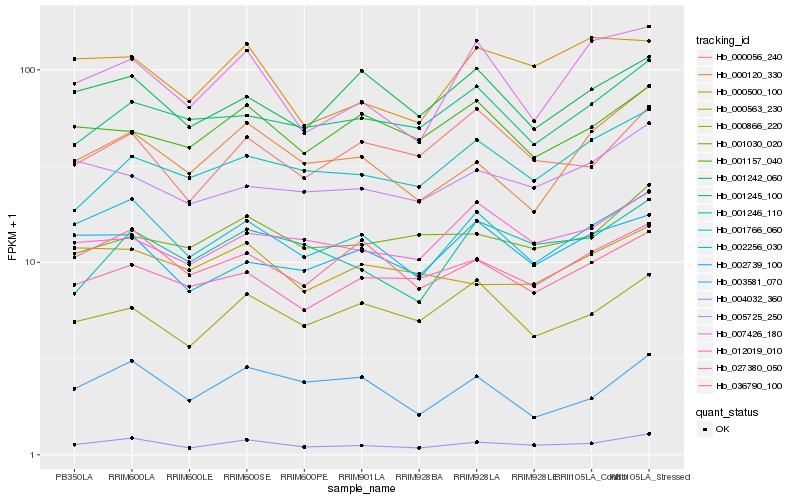
Hb_004032_360 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103403885 [Malus domestica] |
| 2 |
Hb_002256_030 |
0.064570003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000120_330 |
0.0910212139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027380_050 |
0.0965235564 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290-like [Jatropha curcas] |
| 5 |
Hb_001246_110 |
0.0990213245 |
- |
- |
PREDICTED: probable purine permease 11 [Jatropha curcas] |
| 6 |
Hb_000056_240 |
0.0995492174 |
- |
- |
PREDICTED: cullin-3A [Jatropha curcas] |
| 7 |
Hb_001030_020 |
0.0997205031 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 8 |
Hb_036790_100 |
0.0999433166 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |
| 9 |
Hb_000866_220 |
0.1000943283 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 10 |
Hb_007426_180 |
0.1002105598 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 11 |
Hb_012019_010 |
0.1007173115 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP30 [Jatropha curcas] |
| 12 |
Hb_005725_250 |
0.1011150848 |
- |
- |
PREDICTED: uncharacterized protein LOC105630805 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001245_100 |
0.1041762791 |
transcription factor |
TF Family: Alfin-like |
ATP synthase alpha subunit mitochondrial, putative [Ricinus communis] |
| 14 |
Hb_001766_060 |
0.1042383027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000500_100 |
0.1045486556 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 16 |
Hb_001242_060 |
0.1056962292 |
- |
- |
PREDICTED: uncharacterized protein LOC105139508 [Populus euphratica] |
| 17 |
Hb_002739_100 |
0.1058554251 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000563_230 |
0.1066263668 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 19 |
Hb_001157_040 |
0.1067714594 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 20 |
Hb_003581_070 |
0.1072603258 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Malus domestica] |