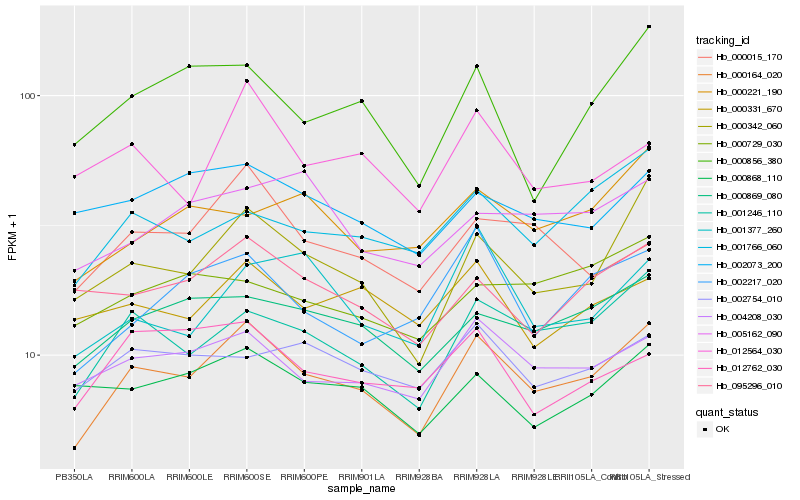
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000164_020 |
0.0 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 2 |
Hb_001246_110 |
0.068446108 |
- |
- |
PREDICTED: probable purine permease 11 [Jatropha curcas] |
| 3 |
Hb_000342_060 |
0.0788229483 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 4 |
Hb_000869_080 |
0.0840190735 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004208_030 |
0.0847288462 |
transcription factor |
TF Family: MYB-related |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 6 |
Hb_001766_060 |
0.0926521613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002217_020 |
0.097692124 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 8 |
Hb_095296_010 |
0.0994398228 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 9 |
Hb_000331_670 |
0.1014659541 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 10 |
Hb_002754_010 |
0.1031061151 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 11 |
Hb_000868_110 |
0.1033379556 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 12 |
Hb_000221_190 |
0.1039537849 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 13 |
Hb_002073_200 |
0.1044047491 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 14 |
Hb_012564_030 |
0.1050771927 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 15 |
Hb_001377_260 |
0.1053371702 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000015_170 |
0.1060212288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000856_380 |
0.1060913334 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 18 |
Hb_000729_030 |
0.1069375706 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 19 |
Hb_012762_030 |
0.1075281488 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 20 |
Hb_005162_090 |
0.1081122241 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |