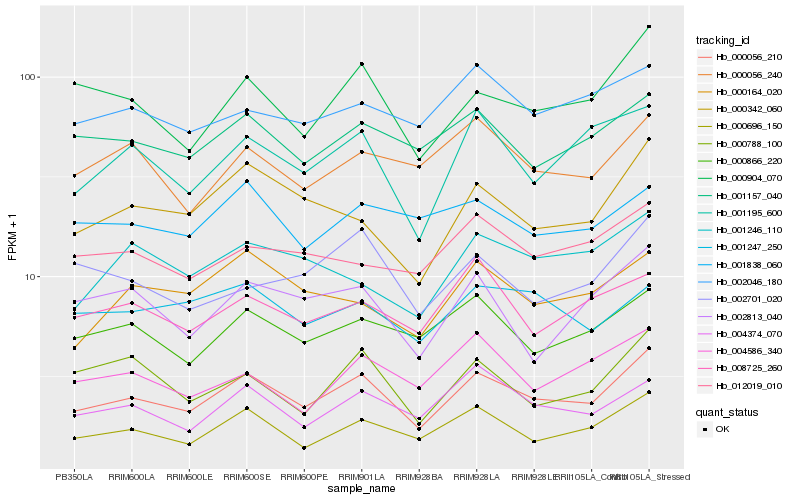
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_210 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26900, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000696_150 |
0.0795079537 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g51320 [Jatropha curcas] |
| 3 |
Hb_000904_070 |
0.0875435022 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001195_600 |
0.0923295895 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000342_060 |
0.0941403996 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 6 |
Hb_000866_220 |
0.0964572878 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 7 |
Hb_004374_070 |
0.096752804 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 8 |
Hb_001157_040 |
0.0993134356 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 9 |
Hb_002813_040 |
0.1065147396 |
- |
- |
PREDICTED: PRA1 family protein G2 [Jatropha curcas] |
| 10 |
Hb_000056_240 |
0.1066677767 |
- |
- |
PREDICTED: cullin-3A [Jatropha curcas] |
| 11 |
Hb_008725_260 |
0.1090155136 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 12 |
Hb_000164_020 |
0.1090650194 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_000788_100 |
0.1092522375 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62890-like [Jatropha curcas] |
| 14 |
Hb_002046_180 |
0.1099331585 |
- |
- |
PREDICTED: uncharacterized protein LOC105641295 [Jatropha curcas] |
| 15 |
Hb_002701_020 |
0.1108504671 |
- |
- |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 16 |
Hb_004586_340 |
0.1113713599 |
- |
- |
PREDICTED: serine/threonine-protein kinase PEPKR2 [Jatropha curcas] |
| 17 |
Hb_012019_010 |
0.1128115821 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP30 [Jatropha curcas] |
| 18 |
Hb_001247_250 |
0.1141666007 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 19 |
Hb_001246_110 |
0.1144662767 |
- |
- |
PREDICTED: probable purine permease 11 [Jatropha curcas] |
| 20 |
Hb_001838_060 |
0.1148609442 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |