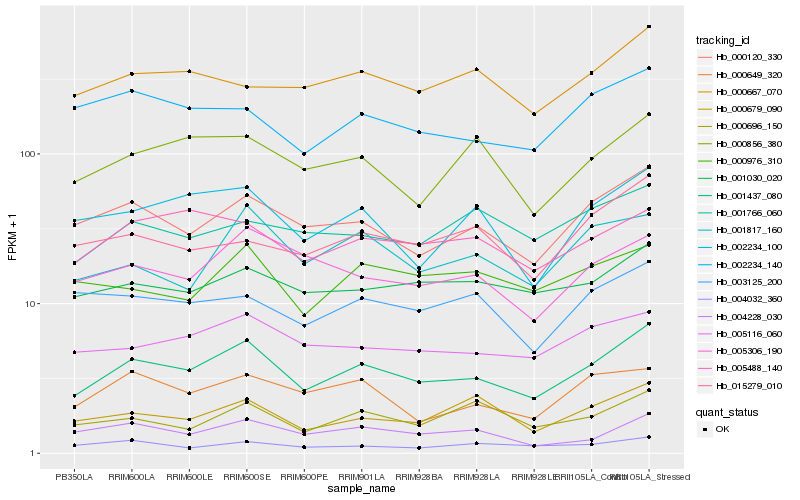
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_080 |
0.0 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 2 |
Hb_000120_330 |
0.0904066583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005116_060 |
0.1114416926 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002234_100 |
0.1123748278 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_001030_020 |
0.1157661244 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 6 |
Hb_000856_380 |
0.1179157342 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 7 |
Hb_005306_190 |
0.1183327057 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 8 |
Hb_000679_090 |
0.1190328512 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 9 |
Hb_005488_140 |
0.1192573125 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 10 |
Hb_001817_160 |
0.1193877028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004228_030 |
0.1203064908 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_002234_140 |
0.1212777683 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 13 |
Hb_000667_070 |
0.1218932888 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 14 |
Hb_000649_320 |
0.1222626776 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 15 |
Hb_015279_010 |
0.1238306658 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 16 |
Hb_000696_150 |
0.1247768601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g51320 [Jatropha curcas] |
| 17 |
Hb_004032_360 |
0.1273567668 |
- |
- |
PREDICTED: uncharacterized protein LOC103403885 [Malus domestica] |
| 18 |
Hb_003125_200 |
0.1279706068 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 19 |
Hb_000976_310 |
0.1282311411 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001766_060 |
0.1288766354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |