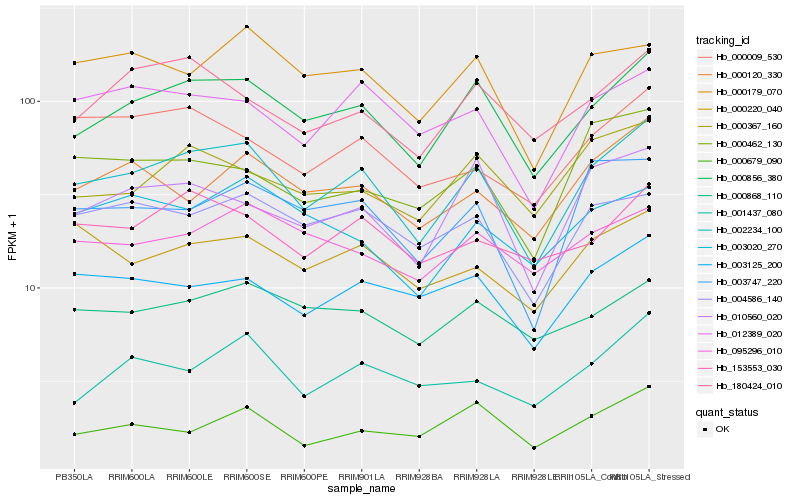
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002234_100 |
0.0 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_000856_380 |
0.0599820089 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 3 |
Hb_012389_020 |
0.0993936658 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 4 |
Hb_004586_140 |
0.1020996937 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 5 |
Hb_003125_200 |
0.1021449326 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 6 |
Hb_003747_220 |
0.1023535476 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_010560_020 |
0.102536589 |
- |
- |
PREDICTED: uncharacterized protein LOC105649267 [Jatropha curcas] |
| 8 |
Hb_000179_070 |
0.1025419905 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 9 |
Hb_000120_330 |
0.1030818485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000220_040 |
0.1065672466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_153553_030 |
0.1071322212 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 12 |
Hb_180424_010 |
0.1074014033 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 13 |
Hb_000367_160 |
0.1092162043 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |
| 14 |
Hb_000009_530 |
0.110142911 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 15 |
Hb_000868_110 |
0.1107030014 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 16 |
Hb_001437_080 |
0.1123748278 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 17 |
Hb_000462_130 |
0.1133023211 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 18 |
Hb_095296_010 |
0.115011989 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 19 |
Hb_000679_090 |
0.1165364741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 20 |
Hb_003020_270 |
0.1168631952 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |