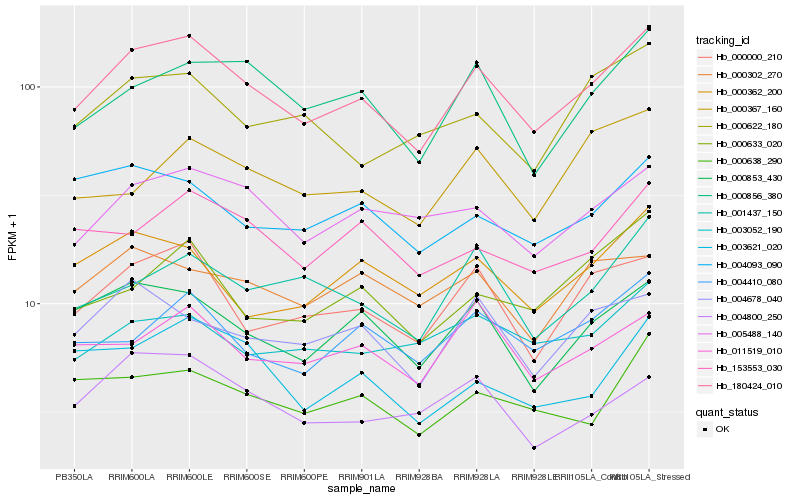
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180424_010 |
0.0 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 2 |
Hb_000000_210 |
0.0790475379 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000853_430 |
0.0830889946 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 4 |
Hb_005488_140 |
0.0848754302 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 5 |
Hb_000856_380 |
0.0870672306 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 6 |
Hb_004410_080 |
0.0943405547 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_004678_040 |
0.0952919015 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 8 |
Hb_011519_010 |
0.0953754633 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 9 |
Hb_153553_030 |
0.0960321418 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 10 |
Hb_001437_150 |
0.0962030771 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 11 |
Hb_004800_250 |
0.0975364476 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 12 |
Hb_000362_200 |
0.0995173889 |
- |
- |
exonuclease family protein [Populus trichocarpa] |
| 13 |
Hb_000638_290 |
0.1001639598 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 14 |
Hb_003052_190 |
0.1017416897 |
- |
- |
PREDICTED: uncharacterized protein LOC105641937 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004093_090 |
0.1023036492 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 16 |
Hb_000302_270 |
0.1026325417 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000622_180 |
0.1027876429 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 18 |
Hb_000633_020 |
0.103144321 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 19 |
Hb_003621_020 |
0.1040052146 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 20 |
Hb_000367_160 |
0.1056081753 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |