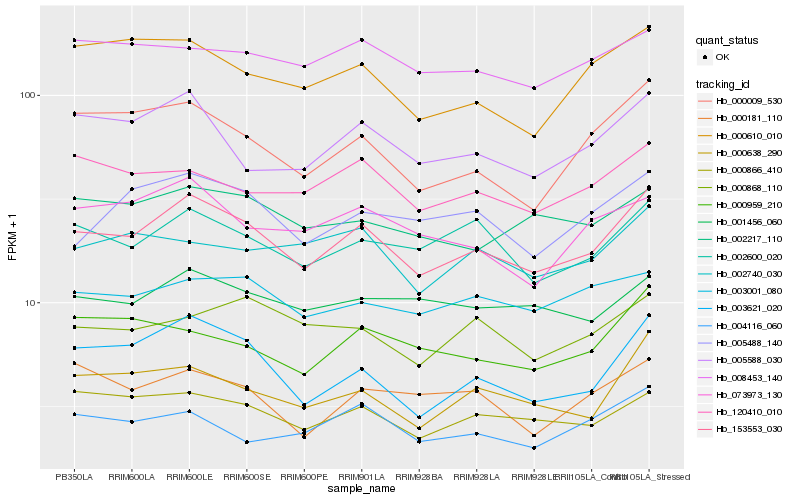
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153553_030 |
0.0 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 2 |
Hb_003621_020 |
0.0748174823 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 3 |
Hb_002600_020 |
0.0762965846 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_005588_030 |
0.0775030913 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 5 |
Hb_000866_410 |
0.0780339916 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 6 |
Hb_000638_290 |
0.0797320345 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 7 |
Hb_000009_530 |
0.0799133809 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 8 |
Hb_005488_140 |
0.0855773037 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 9 |
Hb_120410_010 |
0.0861928653 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 10 |
Hb_000610_010 |
0.086842805 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 11 |
Hb_003001_080 |
0.0890065164 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 12 |
Hb_001456_060 |
0.0891591923 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 13 |
Hb_073973_130 |
0.0895150607 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_002217_110 |
0.0904291683 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 15 |
Hb_008453_140 |
0.0907909775 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 16 |
Hb_000959_210 |
0.0908421274 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 17 |
Hb_002740_030 |
0.0910657636 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004116_060 |
0.0919692161 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 19 |
Hb_000868_110 |
0.0925417281 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 20 |
Hb_000181_110 |
0.0946591874 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |