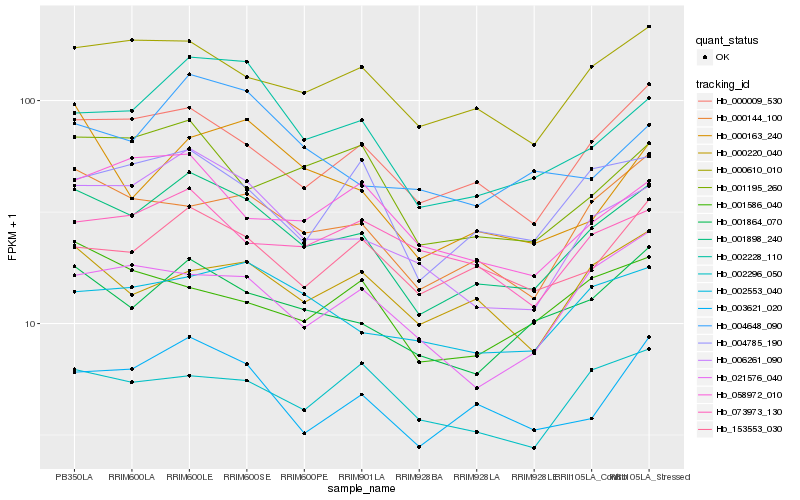
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 2 |
Hb_006261_090 |
0.0761850405 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 3 |
Hb_001864_070 |
0.0790812803 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 4 |
Hb_000009_530 |
0.0814112844 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 5 |
Hb_002228_110 |
0.0848616774 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 6 |
Hb_000610_010 |
0.0852129522 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 7 |
Hb_003621_020 |
0.0874398926 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 8 |
Hb_000144_100 |
0.0884178381 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002553_040 |
0.098049299 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 10 |
Hb_004785_190 |
0.0990136454 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 11 |
Hb_153553_030 |
0.0994387666 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 12 |
Hb_001195_260 |
0.099672832 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 13 |
Hb_000220_040 |
0.1058620591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004648_090 |
0.1077559006 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 15 |
Hb_021576_040 |
0.1078725351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002296_050 |
0.1079726904 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 17 |
Hb_058972_010 |
0.109836972 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001586_040 |
0.1119673578 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 19 |
Hb_000163_240 |
0.1120304129 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |
| 20 |
Hb_073973_130 |
0.1128321736 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |