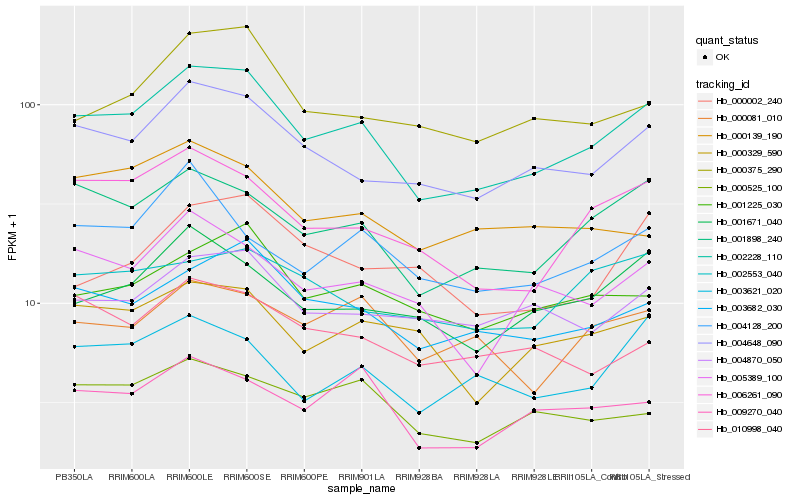
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002228_110 |
0.0 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 2 |
Hb_004648_090 |
0.0792827119 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 3 |
Hb_001898_240 |
0.0848616774 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 4 |
Hb_003682_030 |
0.0858658528 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 5 |
Hb_006261_090 |
0.0924529283 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 6 |
Hb_001671_040 |
0.1006743743 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 7 |
Hb_001225_030 |
0.1008535139 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 8 |
Hb_005389_100 |
0.1020556967 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000375_290 |
0.1033074316 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_004870_050 |
0.1040306701 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 11 |
Hb_000525_100 |
0.1041957698 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 12 |
Hb_000329_590 |
0.1045412935 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_009270_040 |
0.1064388093 |
- |
- |
- |
| 14 |
Hb_000081_010 |
0.1091063984 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 15 |
Hb_003621_020 |
0.1104099567 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 16 |
Hb_010998_040 |
0.1111560397 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 17 |
Hb_004128_200 |
0.1147143363 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 18 |
Hb_002553_040 |
0.1148806581 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 19 |
Hb_000139_190 |
0.1179395366 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000002_240 |
0.1187372758 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |