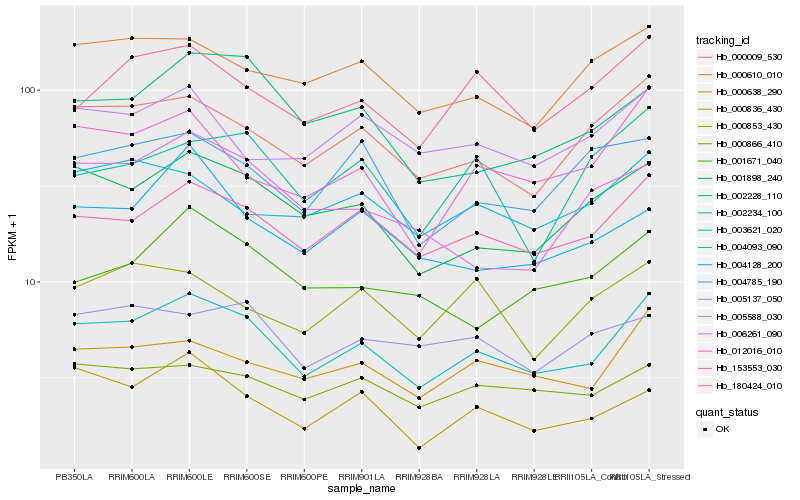
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003621_020 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 2 |
Hb_153553_030 |
0.0748174823 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 3 |
Hb_000009_530 |
0.0859105068 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 4 |
Hb_001898_240 |
0.0874398926 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 5 |
Hb_000638_290 |
0.0894609439 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 6 |
Hb_012016_010 |
0.0988250301 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 7 |
Hb_180424_010 |
0.1040052146 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 8 |
Hb_000866_410 |
0.1051045206 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 9 |
Hb_000610_010 |
0.1065406488 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 10 |
Hb_002228_110 |
0.1104099567 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 11 |
Hb_004093_090 |
0.1131821028 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 12 |
Hb_000836_430 |
0.114640481 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_005588_030 |
0.1159511086 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 14 |
Hb_001671_040 |
0.1160024869 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 15 |
Hb_005137_050 |
0.1163886077 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 16 |
Hb_004785_190 |
0.1176031253 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 17 |
Hb_002234_100 |
0.1188708529 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 18 |
Hb_006261_090 |
0.1205948214 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 19 |
Hb_004128_200 |
0.1209485812 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 20 |
Hb_000853_430 |
0.1210368218 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |