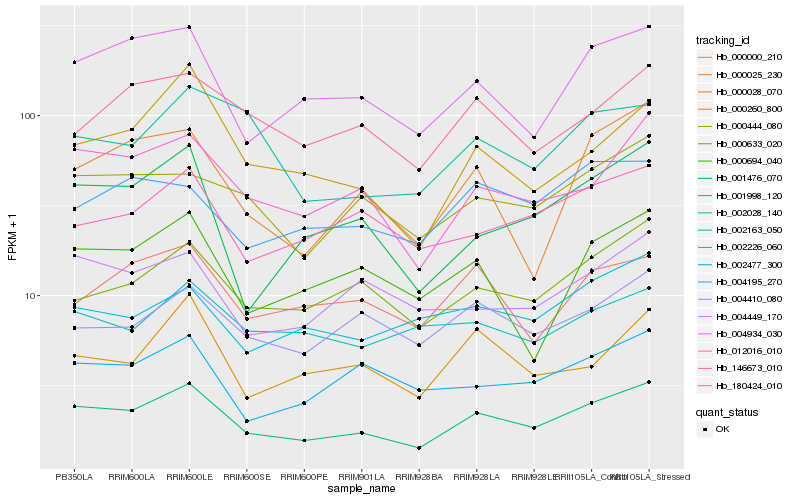
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 2 |
Hb_012016_010 |
0.0859547697 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 3 |
Hb_004934_030 |
0.0946962845 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 4 |
Hb_000633_020 |
0.0986802067 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 5 |
Hb_000444_080 |
0.1025738468 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 6 |
Hb_000260_800 |
0.1094469332 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 7 |
Hb_001476_070 |
0.1102853379 |
- |
- |
MinE protein [Manihot esculenta] |
| 8 |
Hb_004410_080 |
0.1104559021 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_004449_170 |
0.1105341053 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 10 |
Hb_146673_010 |
0.1116595828 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 11 |
Hb_002477_300 |
0.112599687 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_180424_010 |
0.1135053848 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 13 |
Hb_000028_070 |
0.1150921571 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 14 |
Hb_000694_040 |
0.1158140339 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000000_210 |
0.1167960162 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002226_060 |
0.11902704 |
- |
- |
PREDICTED: uncharacterized protein LOC105641377 [Jatropha curcas] |
| 17 |
Hb_004195_270 |
0.1195443201 |
- |
- |
Ubiquitin-conjugating enzyme 16 [Theobroma cacao] |
| 18 |
Hb_002028_140 |
0.1208483634 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 19 |
Hb_000025_230 |
0.1218621951 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |
| 20 |
Hb_002163_050 |
0.1225111921 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |