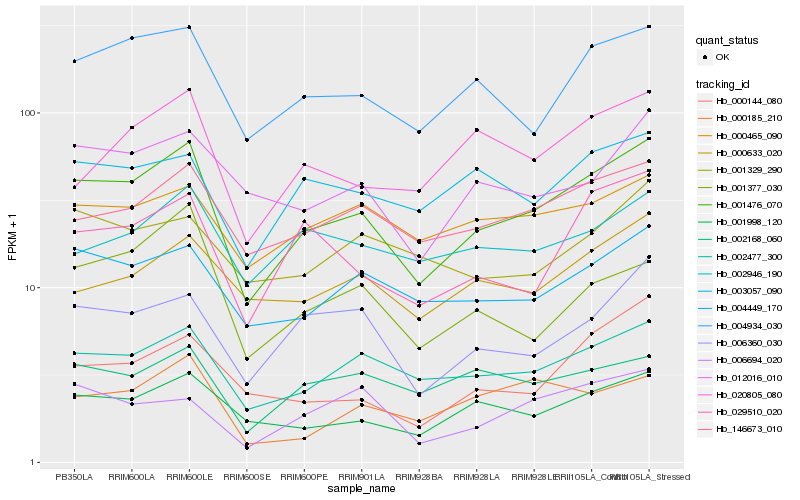
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001476_070 |
0.0 |
- |
- |
MinE protein [Manihot esculenta] |
| 2 |
Hb_002477_300 |
0.102969887 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_004934_030 |
0.1085330084 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 4 |
Hb_001998_120 |
0.1102853379 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 5 |
Hb_004449_170 |
0.1167517191 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 6 |
Hb_146673_010 |
0.1190855219 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 7 |
Hb_006360_030 |
0.1209503646 |
- |
- |
- |
| 8 |
Hb_029510_020 |
0.1253223906 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 9 |
Hb_012016_010 |
0.1278372744 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003057_090 |
0.132933169 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 11 |
Hb_000633_020 |
0.1346889126 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 12 |
Hb_000144_080 |
0.1363540505 |
- |
- |
PREDICTED: uncharacterized protein LOC104609774 [Nelumbo nucifera] |
| 13 |
Hb_020805_080 |
0.1371394615 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 14 |
Hb_000465_090 |
0.1387545385 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 15 |
Hb_002168_060 |
0.1403158608 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_006694_020 |
0.1414607174 |
- |
- |
PREDICTED: auxin response factor 2-like isoform X2 [Nicotiana tomentosiformis] |
| 17 |
Hb_002946_190 |
0.1416835478 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001377_030 |
0.1421140312 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 19 |
Hb_000185_210 |
0.1435114729 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001329_290 |
0.1443842089 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |