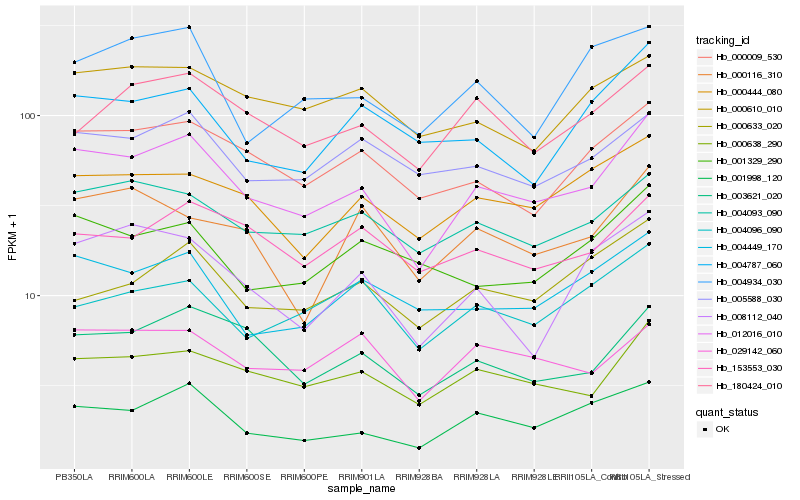
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012016_010 |
0.0 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000638_290 |
0.0852886032 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 3 |
Hb_001998_120 |
0.0859547697 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 4 |
Hb_000009_530 |
0.0959915689 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 5 |
Hb_003621_020 |
0.0988250301 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 6 |
Hb_004093_090 |
0.1003137223 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 7 |
Hb_000444_080 |
0.1025219503 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 8 |
Hb_004449_170 |
0.1057676746 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005588_030 |
0.1107076396 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 10 |
Hb_029142_060 |
0.1121977687 |
- |
- |
PREDICTED: RNA pseudouridine synthase 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_180424_010 |
0.1141985445 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 12 |
Hb_000610_010 |
0.1151978888 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_153553_030 |
0.1164300645 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 14 |
Hb_008112_040 |
0.1164781223 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 15 |
Hb_000633_020 |
0.1179717776 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 16 |
Hb_001329_290 |
0.1185189189 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 17 |
Hb_004787_060 |
0.1205432404 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 18 |
Hb_004096_090 |
0.1205520064 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_004934_030 |
0.1209790309 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 20 |
Hb_000116_310 |
0.1214294816 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |