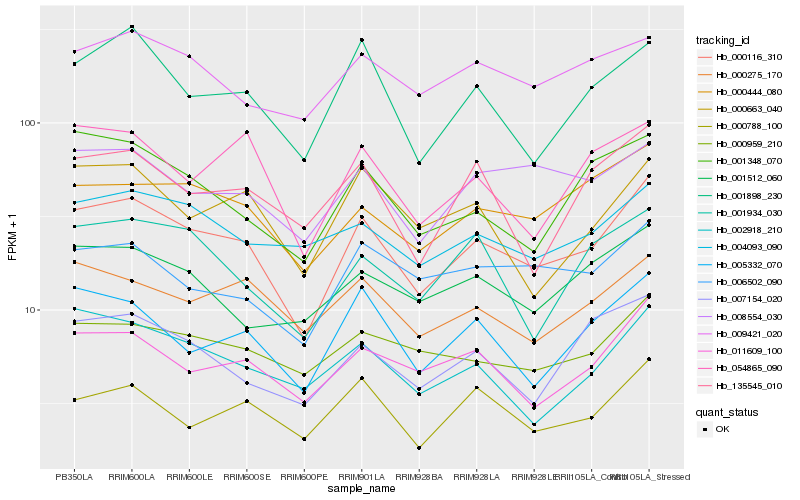
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_310 |
0.0 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |
| 2 |
Hb_000444_080 |
0.0953183932 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 3 |
Hb_011609_100 |
0.095399767 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_005332_070 |
0.0994520764 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |
| 5 |
Hb_001898_230 |
0.1023914771 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000275_170 |
0.1066883179 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 7 |
Hb_000959_210 |
0.1068410695 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 8 |
Hb_008554_030 |
0.107179977 |
- |
- |
PREDICTED: dnaJ homolog 1, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_000788_100 |
0.108127552 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62890-like [Jatropha curcas] |
| 10 |
Hb_001348_070 |
0.1085786464 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_007154_020 |
0.1092646136 |
- |
- |
- |
| 12 |
Hb_006502_090 |
0.1093765669 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 13 |
Hb_054865_090 |
0.1094057959 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 14 |
Hb_002918_210 |
0.1100411438 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 15 |
Hb_000663_040 |
0.1106898343 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004093_090 |
0.1114112539 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 17 |
Hb_001512_060 |
0.1114845611 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001934_030 |
0.1115469228 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 19 |
Hb_135545_010 |
0.1127552975 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_009421_020 |
0.1134145126 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |