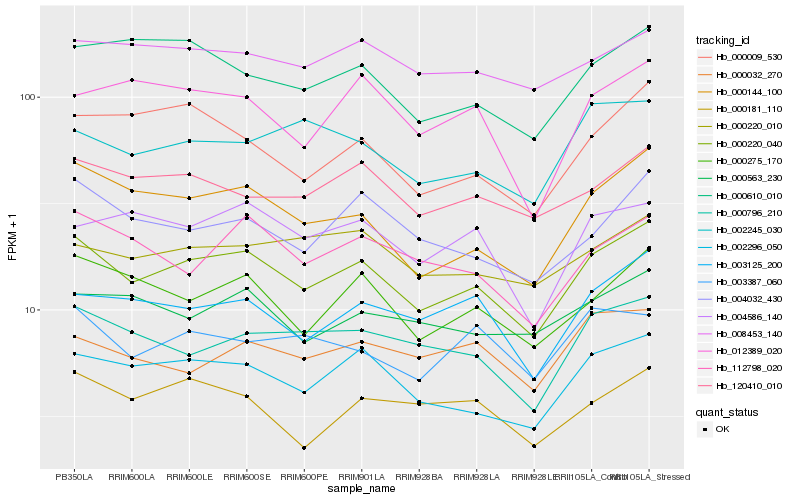
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002296_050 |
0.0663400299 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 3 |
Hb_000144_100 |
0.0689287227 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000275_170 |
0.0723146032 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 5 |
Hb_003125_200 |
0.0794227327 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 6 |
Hb_004032_430 |
0.0809043699 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 7 |
Hb_000796_210 |
0.0815299641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_120410_010 |
0.082789186 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 9 |
Hb_112798_020 |
0.0859789033 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 10 |
Hb_000610_010 |
0.0863160152 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 11 |
Hb_000009_530 |
0.0867897698 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 12 |
Hb_000563_230 |
0.0892370194 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 13 |
Hb_004586_140 |
0.0893104116 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 14 |
Hb_000220_010 |
0.0896434605 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 15 |
Hb_002245_030 |
0.0904563806 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_008453_140 |
0.0911069373 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 17 |
Hb_000181_110 |
0.0922220917 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 18 |
Hb_003387_060 |
0.0929092978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012389_020 |
0.0934272371 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 20 |
Hb_000032_270 |
0.0948580357 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13 [Jatropha curcas] |