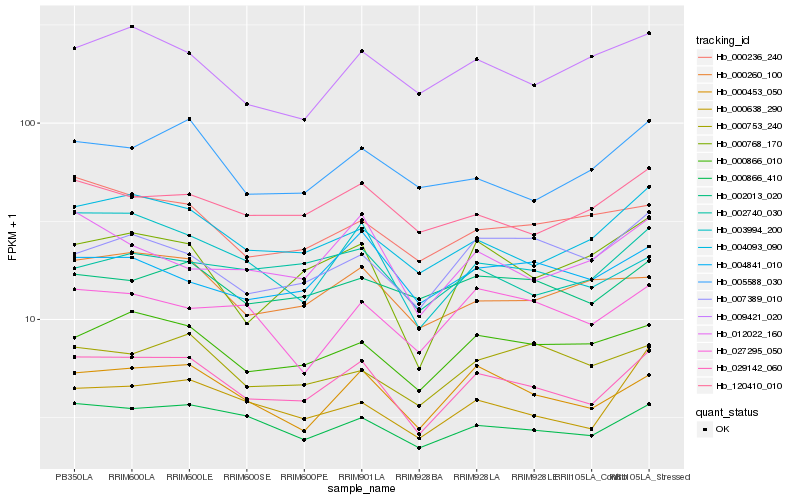
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029142_060 |
0.0 |
- |
- |
PREDICTED: RNA pseudouridine synthase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000453_050 |
0.0646479443 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 3 |
Hb_000866_410 |
0.0724224087 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 4 |
Hb_000866_010 |
0.0753764119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004093_090 |
0.07674435 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 6 |
Hb_000260_100 |
0.0854492902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003994_200 |
0.0880186808 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 8 |
Hb_000638_290 |
0.0893714267 |
- |
- |
PREDICTED: 5' exonuclease Apollo [Jatropha curcas] |
| 9 |
Hb_120410_010 |
0.0904292131 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 10 |
Hb_000768_170 |
0.0919132822 |
- |
- |
hypothetical protein JCGZ_02624 [Jatropha curcas] |
| 11 |
Hb_000236_240 |
0.0924628234 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 12 |
Hb_000753_240 |
0.0933564308 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 13 |
Hb_005588_030 |
0.0938688377 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 14 |
Hb_002013_020 |
0.0941308183 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 15 |
Hb_009421_020 |
0.0947629298 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 16 |
Hb_007389_010 |
0.0951278705 |
- |
- |
PREDICTED: THO complex subunit 7A [Jatropha curcas] |
| 17 |
Hb_012022_160 |
0.0954541555 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 18 |
Hb_002740_030 |
0.0954927809 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004841_010 |
0.0979400328 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 20 |
Hb_027295_050 |
0.0981144866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48730, chloroplastic [Jatropha curcas] |