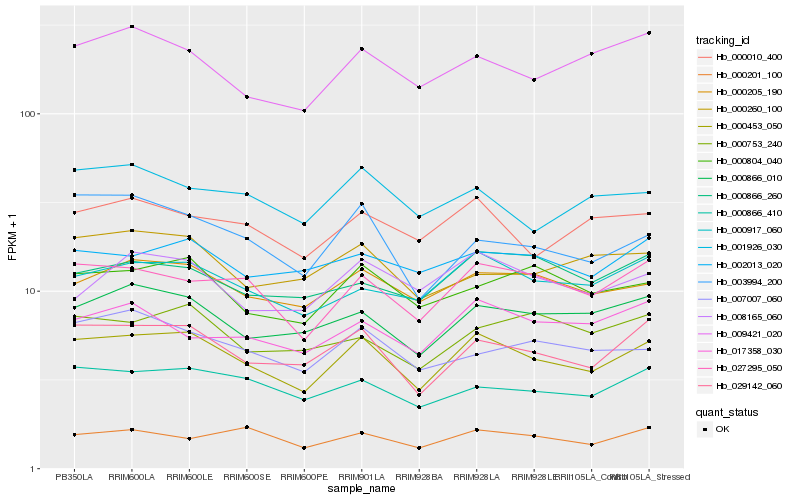
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_050 |
0.0 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 2 |
Hb_029142_060 |
0.0646479443 |
- |
- |
PREDICTED: RNA pseudouridine synthase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_027295_050 |
0.0719572489 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48730, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000917_060 |
0.0807277416 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 5 |
Hb_000205_190 |
0.0813072391 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000866_010 |
0.0821874228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000866_410 |
0.0835957967 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 8 |
Hb_000804_040 |
0.0888288251 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000010_400 |
0.0911383835 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 10 |
Hb_009421_020 |
0.0912607354 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 11 |
Hb_003994_200 |
0.0927282093 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 12 |
Hb_008165_060 |
0.0955544451 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 13 |
Hb_002013_020 |
0.0968684123 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 14 |
Hb_001926_030 |
0.0969415673 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 15 |
Hb_007007_060 |
0.0981089637 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_017358_030 |
0.0987621226 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000201_100 |
0.0991834903 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 18 |
Hb_000753_240 |
0.0999168203 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 19 |
Hb_000866_260 |
0.1004208301 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000260_100 |
0.1006439982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |