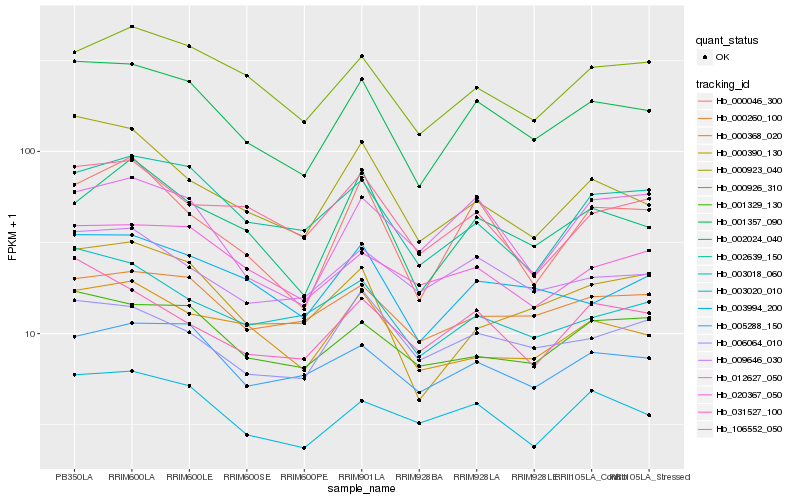
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_090 |
0.0 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 2 |
Hb_003994_200 |
0.0823078149 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 3 |
Hb_003020_010 |
0.0874025059 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 4 |
Hb_000926_310 |
0.0880493955 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 5 |
Hb_000046_300 |
0.088639603 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 6 |
Hb_002024_040 |
0.0913259634 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 7 |
Hb_001329_130 |
0.0914532524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005288_150 |
0.0923741573 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_000923_040 |
0.0933118713 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 10 |
Hb_002639_150 |
0.094275725 |
- |
- |
- |
| 11 |
Hb_009646_030 |
0.0944479525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000368_020 |
0.0951089409 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_031527_100 |
0.0966522415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000390_130 |
0.0966871522 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 15 |
Hb_000260_100 |
0.0980216431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003018_060 |
0.0984593988 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 17 |
Hb_006064_010 |
0.0993740465 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 4 [Jatropha curcas] |
| 18 |
Hb_020367_050 |
0.0998362829 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_012627_050 |
0.100361981 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_106552_050 |
0.1020825999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |