| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
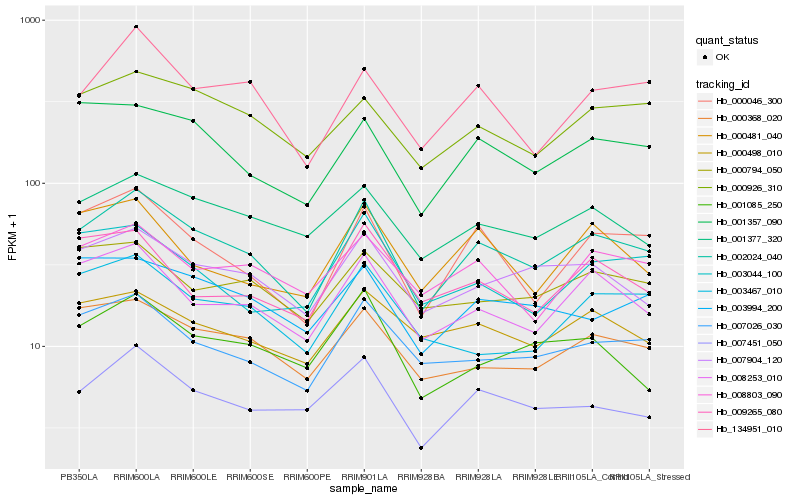
Hb_002024_040 |
0.0 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 2 |
Hb_000046_300 |
0.0848241292 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 3 |
Hb_008253_010 |
0.0849975463 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 4 |
Hb_007026_030 |
0.0874115503 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 5 |
Hb_000368_020 |
0.0908263656 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_007904_120 |
0.0910221947 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001357_090 |
0.0913259634 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 8 |
Hb_007451_050 |
0.0913464583 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001377_320 |
0.093550423 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 10 |
Hb_000926_310 |
0.0953310522 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 11 |
Hb_134951_010 |
0.0987113502 |
- |
- |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 12 |
Hb_000498_010 |
0.1019658535 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_008803_090 |
0.1022438583 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 14 |
Hb_003994_200 |
0.1067990444 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 15 |
Hb_000794_050 |
0.1082516352 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 16 |
Hb_009265_080 |
0.1098182031 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 17 |
Hb_001085_250 |
0.1101159206 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL isoform X2 [Jatropha curcas] |
| 18 |
Hb_000481_040 |
0.1102267021 |
- |
- |
developmentally regulated GTP-binding protein, putative [Ricinus communis] |
| 19 |
Hb_003044_100 |
0.1116119095 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 20 |
Hb_003467_010 |
0.1122412239 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |