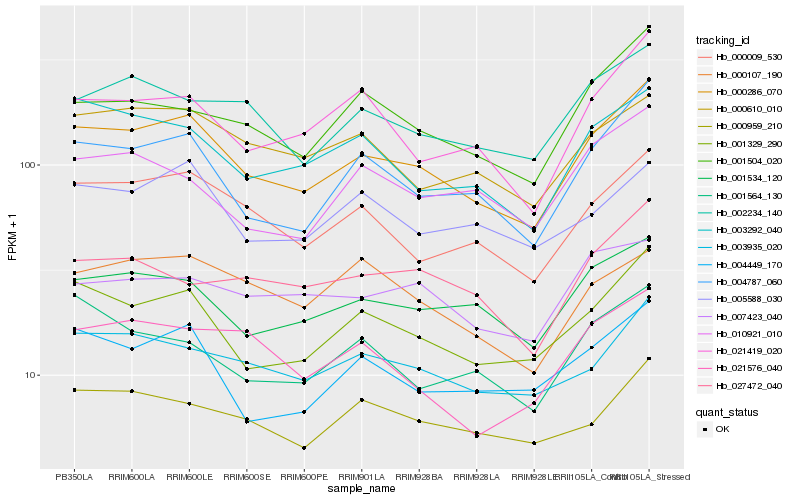
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000286_070 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 2 |
Hb_001329_290 |
0.0585582128 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 3 |
Hb_004787_060 |
0.0648759645 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 4 |
Hb_000009_530 |
0.06698475 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 5 |
Hb_001534_120 |
0.0761162798 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 6 |
Hb_003292_040 |
0.0774320033 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 7 |
Hb_003935_020 |
0.0779507177 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001504_020 |
0.0792316489 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 9 |
Hb_002234_140 |
0.0799203933 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 10 |
Hb_004449_170 |
0.0802564039 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021419_020 |
0.0812858906 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 12 |
Hb_000610_010 |
0.0835073966 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_000959_210 |
0.0874840447 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 14 |
Hb_001564_130 |
0.0899851885 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_005588_030 |
0.090054027 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 16 |
Hb_021576_040 |
0.0915824538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010921_010 |
0.0932473062 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 18 |
Hb_027472_040 |
0.0937696556 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 19 |
Hb_007423_040 |
0.0957564122 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000107_190 |
0.0963037651 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |