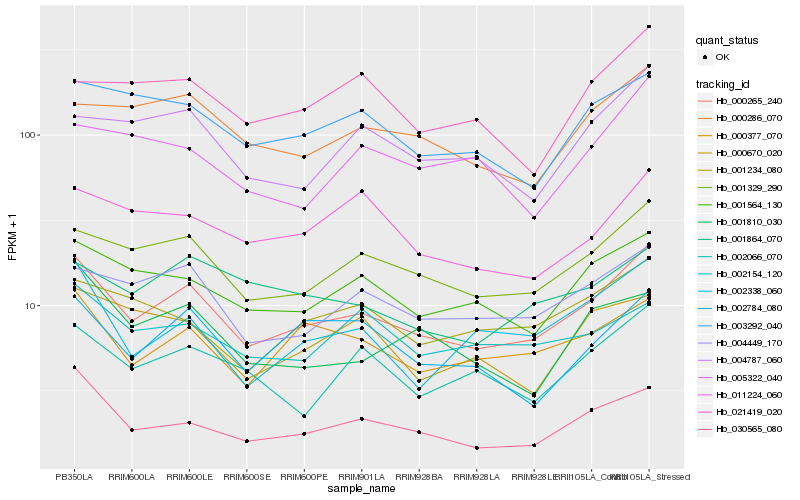
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000265_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643590 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001329_290 |
0.0815708839 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 3 |
Hb_001564_130 |
0.0987360413 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_004449_170 |
0.0989503527 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000377_070 |
0.1019300869 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 [Jatropha curcas] |
| 6 |
Hb_030565_080 |
0.1027522831 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 7 |
Hb_003292_040 |
0.1134912162 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 8 |
Hb_000286_070 |
0.1139409091 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 9 |
Hb_002338_060 |
0.1144085603 |
- |
- |
2Fe-2S ferredoxin [Glycine soja] |
| 10 |
Hb_002784_080 |
0.1155628006 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF3-like [Jatropha curcas] |
| 11 |
Hb_002066_070 |
0.1168095781 |
- |
- |
PREDICTED: uncharacterized protein LOC105631043 [Jatropha curcas] |
| 12 |
Hb_004787_060 |
0.1170833943 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 13 |
Hb_001234_080 |
0.1176221167 |
- |
- |
PREDICTED: eyes absent homolog 2-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_011224_060 |
0.1223548539 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 15 |
Hb_002154_120 |
0.1240815734 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 16 |
Hb_001864_070 |
0.1248730937 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 17 |
Hb_021419_020 |
0.12992165 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 18 |
Hb_001810_030 |
0.1299541244 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_005322_040 |
0.1304230752 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 20 |
Hb_000670_020 |
0.1306137617 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |