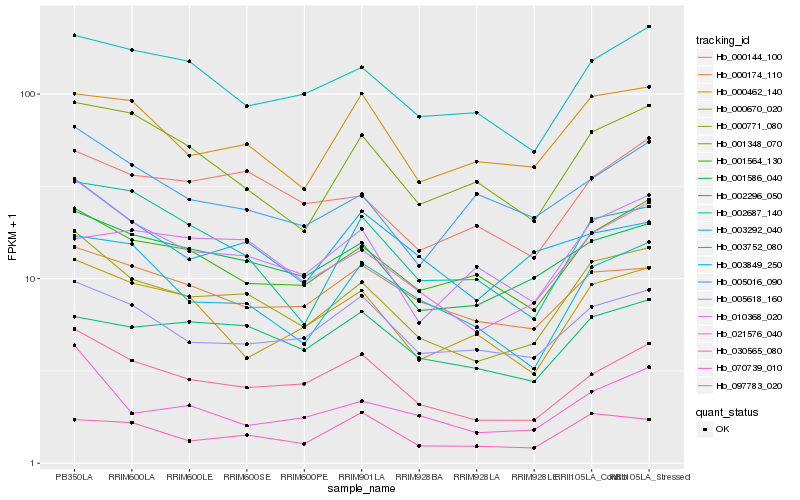
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000771_080 |
0.0 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 2 |
Hb_010368_020 |
0.0806896991 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 3 |
Hb_097783_020 |
0.0831149353 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 4 |
Hb_001586_040 |
0.0882448775 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 5 |
Hb_001564_130 |
0.0980062964 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_005618_160 |
0.1038903132 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 7 |
Hb_000144_100 |
0.1057926586 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_003292_040 |
0.1062039528 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 9 |
Hb_001348_070 |
0.1107640436 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_030565_080 |
0.1109621254 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 11 |
Hb_003752_080 |
0.1114925569 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000462_140 |
0.1127704069 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 13 |
Hb_003849_250 |
0.1148674041 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 14 |
Hb_000670_020 |
0.1152486078 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 15 |
Hb_000174_110 |
0.1156895429 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 16 |
Hb_021576_040 |
0.1174794176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002687_140 |
0.1179308118 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 18 |
Hb_070739_010 |
0.1200418494 |
- |
- |
PREDICTED: probable aminotransferase ACS12 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005016_090 |
0.1205183676 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 20 |
Hb_002296_050 |
0.1216243145 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |