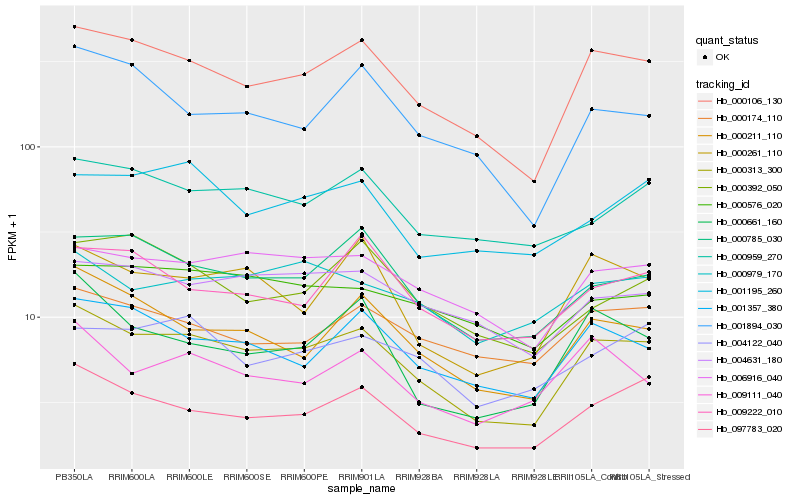
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_300 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Jatropha curcas] |
| 2 |
Hb_000106_130 |
0.0569807035 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_097783_020 |
0.0937509151 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 4 |
Hb_000211_110 |
0.0941245153 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_001357_380 |
0.0970253745 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_000785_030 |
0.0983741787 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 7 |
Hb_006916_040 |
0.1001722132 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 8 |
Hb_000261_110 |
0.1049710128 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_001894_030 |
0.1115114984 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 10 |
Hb_004631_180 |
0.1116075762 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 11 |
Hb_000959_270 |
0.1118165468 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 12 |
Hb_009111_040 |
0.1139003166 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 13 |
Hb_000661_160 |
0.1142398289 |
- |
- |
hexokinase [Manihot esculenta] |
| 14 |
Hb_000174_110 |
0.1157073049 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 15 |
Hb_009222_010 |
0.1171033453 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_000979_170 |
0.1182348567 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 17 |
Hb_004122_040 |
0.1190008825 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 18 |
Hb_001195_260 |
0.1192714895 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 19 |
Hb_000576_020 |
0.1207099614 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 20 |
Hb_000392_050 |
0.1215851742 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |