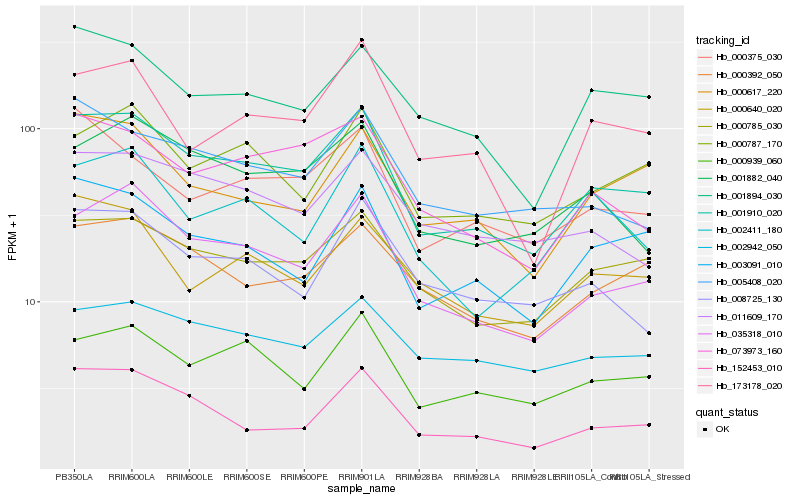
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001910_020 |
0.0 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_035318_010 |
0.0712206946 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 3 |
Hb_000785_030 |
0.0810034189 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 4 |
Hb_152453_010 |
0.0889198133 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003091_010 |
0.0924655832 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 6 |
Hb_073973_160 |
0.0926327599 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_001882_040 |
0.0947030865 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 8 |
Hb_000939_060 |
0.0948889595 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 9 |
Hb_000392_050 |
0.096562715 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001894_030 |
0.0974612285 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 11 |
Hb_000787_170 |
0.0987096063 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 12 |
Hb_005408_020 |
0.10180329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011609_170 |
0.1047349443 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 14 |
Hb_000375_030 |
0.1048182391 |
- |
- |
- |
| 15 |
Hb_000617_220 |
0.1059193913 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 16 |
Hb_002942_050 |
0.1072727966 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_173178_020 |
0.1089540651 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000640_020 |
0.1090129965 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 19 |
Hb_002411_180 |
0.1097731839 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008725_130 |
0.1100705451 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |