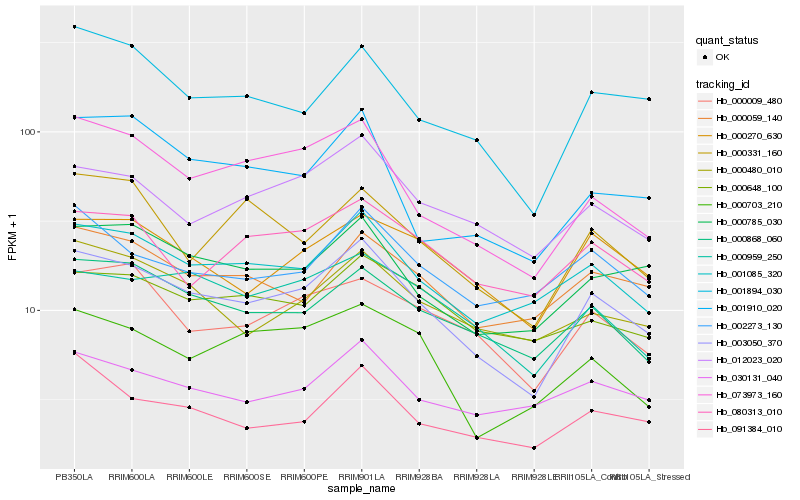
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_370 |
0.0 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 2 |
Hb_001085_320 |
0.0855622393 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_073973_160 |
0.0880240293 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 4 |
Hb_000868_060 |
0.0943307712 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 5 |
Hb_000785_030 |
0.095562625 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 6 |
Hb_001894_030 |
0.0974011475 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 7 |
Hb_000959_250 |
0.0993819088 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 8 |
Hb_012023_020 |
0.1001150795 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 9 |
Hb_000009_480 |
0.1004968596 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 10 |
Hb_000270_630 |
0.1046428697 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 11 |
Hb_000059_140 |
0.1063890446 |
- |
- |
rnase h, putative [Ricinus communis] |
| 12 |
Hb_000480_010 |
0.1067769035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_091384_010 |
0.1091756859 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 14 |
Hb_002273_130 |
0.11040565 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 15 |
Hb_001910_020 |
0.1105528731 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_030131_040 |
0.1107674511 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 17 |
Hb_080313_010 |
0.111149211 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 18 |
Hb_000703_210 |
0.1114673874 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 19 |
Hb_000331_160 |
0.1119976891 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 20 |
Hb_000648_100 |
0.1124619732 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |